VAMP (vesicle-associated membrane protein)-associated protein B and C
ZFIN 
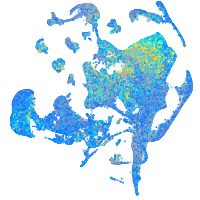
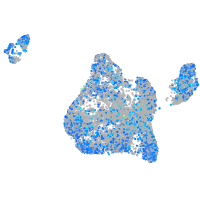
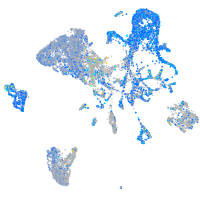
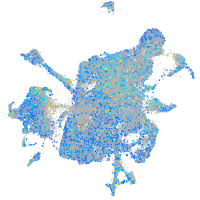
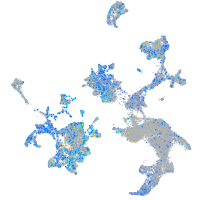
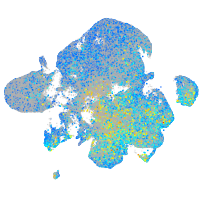
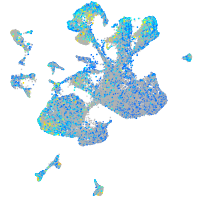
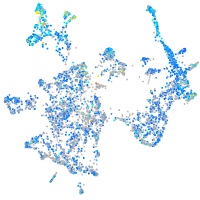
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gnb1a | 0.197 | cx43.4 | -0.125 |
| ywhag2 | 0.194 | hspb1 | -0.118 |
| calm1a | 0.190 | lig1 | -0.117 |
| sncb | 0.183 | mki67 | -0.116 |
| stmn2a | 0.183 | stmn1a | -0.109 |
| rab6bb | 0.182 | apoc1 | -0.107 |
| zgc:65894 | 0.181 | si:dkey-66i24.9 | -0.107 |
| gng3 | 0.177 | si:ch211-152c2.3 | -0.106 |
| atp6v1g1 | 0.175 | chaf1a | -0.105 |
| snap25a | 0.173 | dkc1 | -0.105 |
| atp6v1e1b | 0.171 | zmp:0000000624 | -0.102 |
| mllt11 | 0.170 | anp32b | -0.101 |
| stxbp1a | 0.170 | msna | -0.101 |
| vamp2 | 0.168 | nop58 | -0.101 |
| ywhah | 0.166 | pou5f3 | -0.100 |
| stx1b | 0.164 | tpx2 | -0.100 |
| tpi1b | 0.161 | hnrnpa1b | -0.099 |
| hist2h2l | 0.160 | stm | -0.099 |
| atp6v0cb | 0.159 | zgc:110425 | -0.099 |
| eno2 | 0.159 | fbl | -0.098 |
| gap43 | 0.158 | mibp | -0.098 |
| ldhba | 0.158 | dek | -0.097 |
| atp5if1b | 0.156 | nasp | -0.097 |
| rtn1b | 0.156 | ccnb1 | -0.096 |
| elavl4 | 0.155 | pcna | -0.096 |
| map1aa | 0.155 | akap12b | -0.095 |
| calm3a | 0.155 | dlgap5 | -0.095 |
| calm2a | 0.154 | polr3gla | -0.095 |
| gapdhs | 0.154 | ube2c | -0.095 |
| calm1b | 0.153 | cdca8 | -0.094 |
| ccni | 0.153 | npm1a | -0.094 |
| gabarapl2 | 0.152 | ccna2 | -0.093 |
| rnasekb | 0.152 | COX7A2 | -0.093 |
| zgc:153426 | 0.151 | sp5l | -0.093 |
| ndufa4 | 0.150 | ccnd1 | -0.092 |