ubiquitin specific peptidase 9
ZFIN 
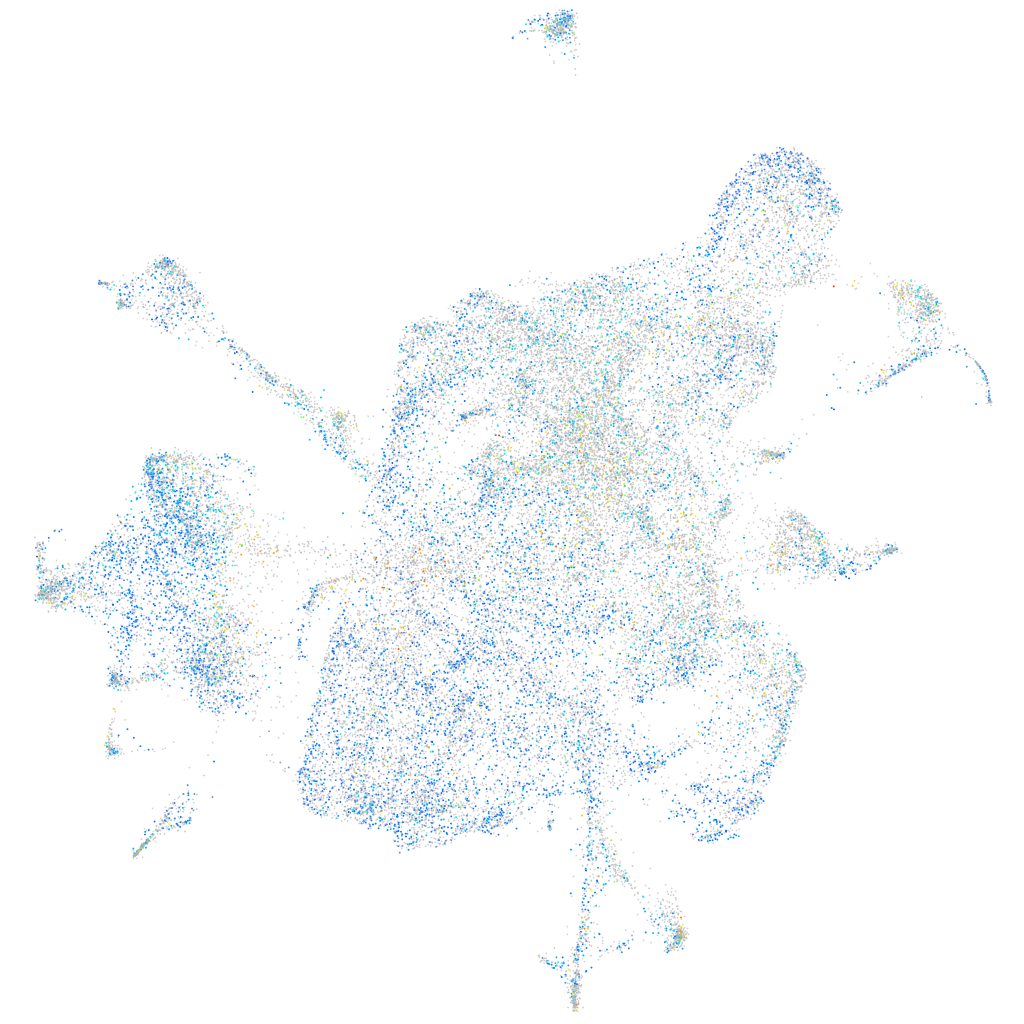
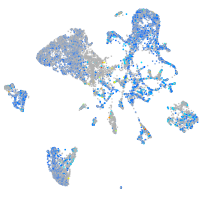
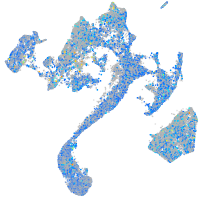
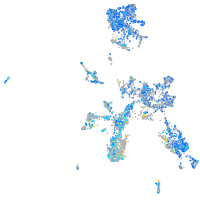
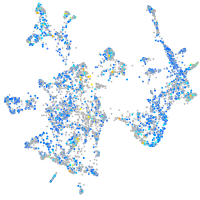
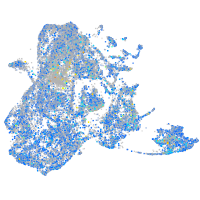
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| marcksb | 0.078 | si:dkey-151g10.6 | -0.053 |
| tuba1c | 0.076 | rpl39 | -0.050 |
| hnrnpa0a | 0.073 | rplp1 | -0.050 |
| elavl3 | 0.073 | rpl37 | -0.046 |
| rtn1b | 0.072 | rps15a | -0.044 |
| ctnnb1 | 0.071 | cebpd | -0.043 |
| mt-co1 | 0.070 | rps29 | -0.041 |
| tuba1a | 0.070 | rpl38 | -0.039 |
| mt-cyb | 0.069 | rpl26 | -0.039 |
| ywhaqb | 0.069 | rps24 | -0.039 |
| hmgb3a | 0.068 | pvalb2 | -0.038 |
| cfl1 | 0.068 | pvalb1 | -0.036 |
| gnb1b | 0.067 | rps28 | -0.035 |
| tubb5 | 0.067 | actc1b | -0.035 |
| stmn1b | 0.066 | rps21 | -0.035 |
| hnrnpa1a | 0.066 | rpl35 | -0.034 |
| gng3 | 0.066 | rpl36a | -0.034 |
| cd99l2 | 0.065 | rpl27 | -0.034 |
| zc4h2 | 0.065 | apoa1b | -0.033 |
| nucks1a | 0.065 | rbp4 | -0.033 |
| nova2 | 0.064 | rps27a | -0.033 |
| igf2bp1 | 0.064 | rpl29 | -0.032 |
| syncrip | 0.064 | rps23 | -0.032 |
| si:ch211-288g17.3 | 0.064 | apoa2 | -0.032 |
| tubb2b | 0.064 | rpl35a | -0.031 |
| si:dkey-276j7.1 | 0.063 | rps19 | -0.031 |
| cirbpa | 0.063 | rpl22 | -0.031 |
| ammecr1 | 0.063 | hpdb | -0.030 |
| pfn2 | 0.062 | zgc:153704 | -0.030 |
| hnrnpub | 0.062 | rpl23 | -0.030 |
| mt-co2 | 0.062 | eef1da | -0.030 |
| hnrnpa0b | 0.062 | rpl19 | -0.030 |
| rtn1a | 0.062 | rps7 | -0.029 |
| tardbp | 0.061 | rpl10a | -0.029 |
| kdm1a | 0.061 | rps26l | -0.029 |