uridine-cytidine kinase 2a
ZFIN 
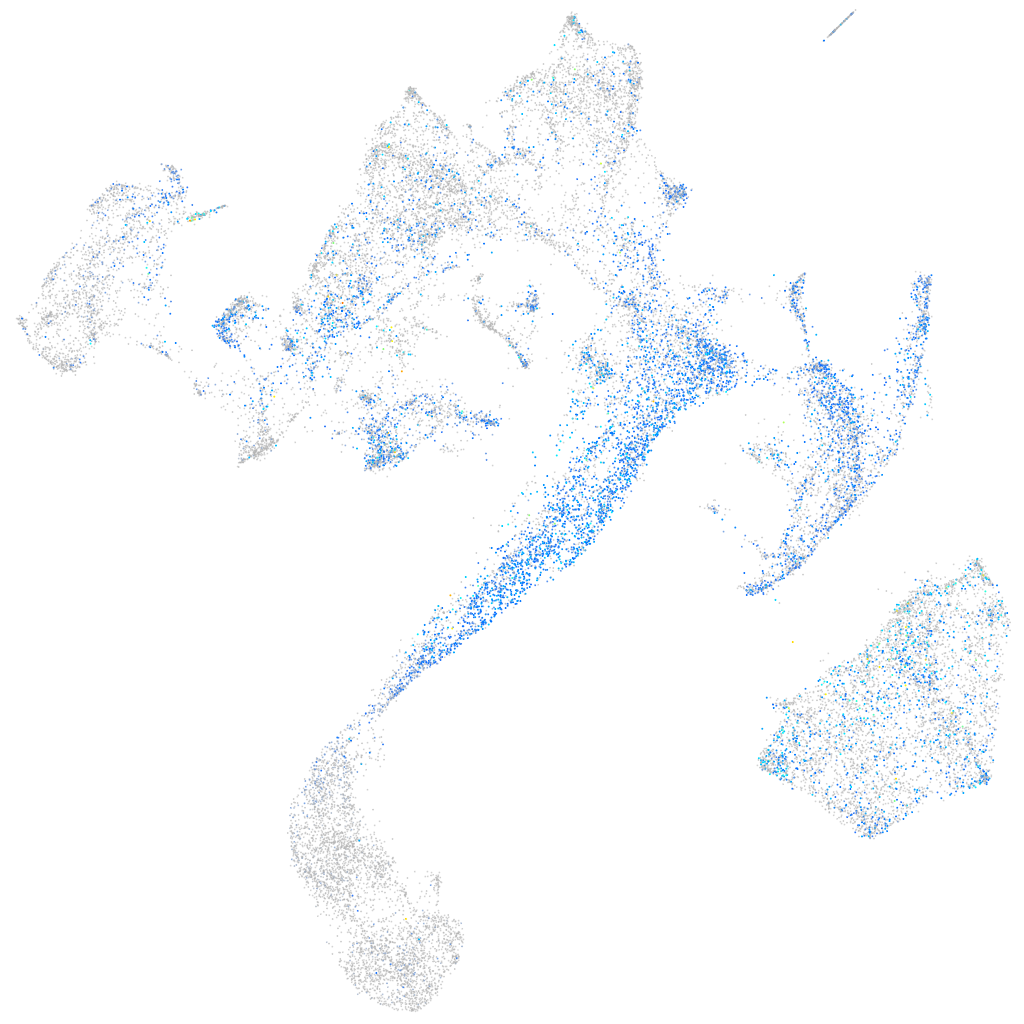
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| snu13b | 0.279 | si:dkey-16p21.8 | -0.217 |
| mafbb | 0.277 | pvalb1 | -0.204 |
| npm1a | 0.273 | bhmt | -0.200 |
| cnbpa | 0.264 | pvalb2 | -0.187 |
| ssb | 0.262 | si:ch211-255p10.3 | -0.184 |
| nhp2 | 0.261 | pkmb | -0.182 |
| jam2a | 0.259 | tnnt3b | -0.181 |
| ddx39ab | 0.258 | actn3a | -0.180 |
| myog | 0.253 | XLOC-025819 | -0.179 |
| nop10 | 0.250 | prx | -0.179 |
| hsp90aa1.1 | 0.250 | casq1b | -0.177 |
| rpl7l1 | 0.249 | si:ch73-367p23.2 | -0.176 |
| btf3l4 | 0.249 | smyd1a | -0.175 |
| nifk | 0.248 | XLOC-006515 | -0.174 |
| c1qbp | 0.246 | pgam2 | -0.174 |
| cct7 | 0.245 | myom1a | -0.172 |
| erh | 0.243 | dhrs7cb | -0.171 |
| mrto4 | 0.243 | XLOC-005350 | -0.171 |
| setb | 0.240 | si:ch211-266g18.10 | -0.170 |
| bccip | 0.240 | eno3 | -0.169 |
| fam107b | 0.239 | XLOC-001975 | -0.169 |
| emg1 | 0.236 | eef2l2 | -0.168 |
| zgc:92429 | 0.235 | tnni2a.4 | -0.168 |
| tomm40l | 0.234 | slc25a4 | -0.168 |
| aktip | 0.234 | CABZ01061524.1 | -0.165 |
| ptges3b | 0.234 | tmod4 | -0.162 |
| serbp1a | 0.234 | ckmt2b | -0.161 |
| nop58 | 0.234 | ndrg2 | -0.161 |
| npm3 | 0.233 | mylpfb | -0.161 |
| cct6a | 0.232 | CR855257.1 | -0.160 |
| pbdc1 | 0.232 | cavin4a | -0.160 |
| snrpd2 | 0.232 | ryr3 | -0.158 |
| zbtb18 | 0.231 | neb | -0.158 |
| mss51 | 0.231 | casq1a | -0.157 |
| tomm20a | 0.229 | glud1b | -0.156 |