UBX domain protein 8
ZFIN 
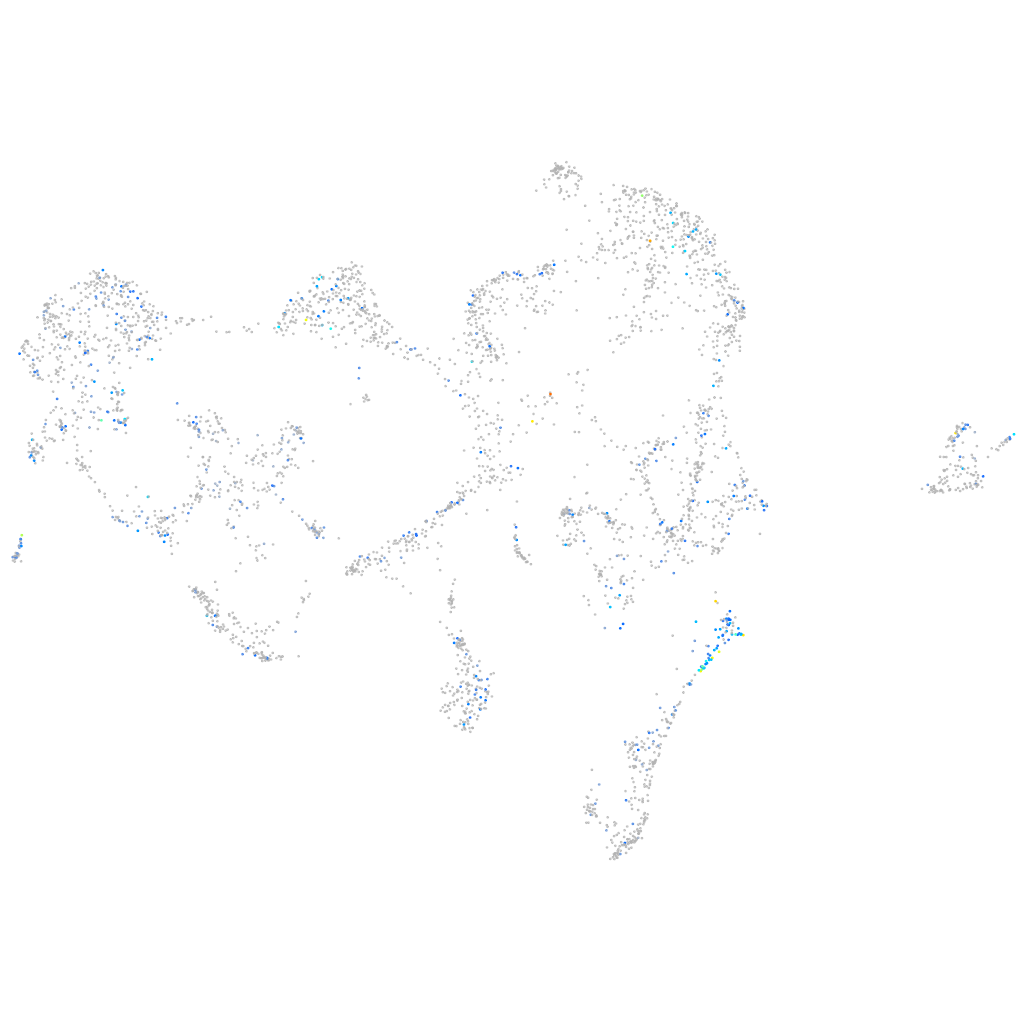
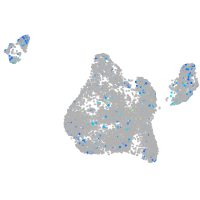
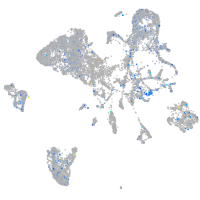
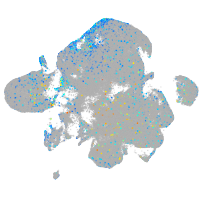
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| BX927258.1 | 0.435 | icn | -0.149 |
| XLOC-032526 | 0.425 | rplp2 | -0.105 |
| XLOC-039121 | 0.420 | krt91 | -0.104 |
| hspb1 | 0.345 | eef1da | -0.095 |
| apoc1 | 0.337 | rpl38 | -0.078 |
| pros1 | 0.311 | pvalb2 | -0.076 |
| XLOC-001603 | 0.304 | pvalb1 | -0.075 |
| CABZ01075068.1 | 0.251 | gapdhs | -0.075 |
| nr6a1a | 0.242 | rps29 | -0.074 |
| CR847906.1 | 0.239 | nme2b.1 | -0.074 |
| nme2a | 0.233 | rpl31 | -0.071 |
| CABZ01038709.1 | 0.230 | adh5 | -0.070 |
| LOC101884747 | 0.230 | agr2 | -0.069 |
| si:dkey-27i16.2 | 0.219 | si:dkey-65b12.6 | -0.065 |
| nid2a | 0.214 | sytl2b | -0.064 |
| LOC103910859 | 0.211 | s100v2 | -0.064 |
| apoeb | 0.208 | rpl14 | -0.064 |
| sfrp5 | 0.201 | fbp1a | -0.062 |
| XLOC-028299 | 0.198 | sdr16c5b | -0.061 |
| arhgef25b | 0.195 | ahcy | -0.061 |
| inaa | 0.195 | crip1 | -0.060 |
| cdx4 | 0.194 | BX000438.2 | -0.060 |
| gata2a | 0.192 | rpl34 | -0.060 |
| XLOC-001964 | 0.187 | rpl37 | -0.060 |
| akap12b | 0.186 | muc5.1 | -0.059 |
| inka1b | 0.186 | calm1b | -0.059 |
| alcamb | 0.186 | mt-co2 | -0.059 |
| zp2.1 | 0.185 | zgc:158463 | -0.058 |
| gfi1ab | 0.182 | COX3 | -0.058 |
| shroom4 | 0.181 | cracr2b | -0.058 |
| XLOC-005432 | 0.180 | cd63 | -0.057 |
| egln1b | 0.178 | atp2a3 | -0.057 |
| cdh13 | 0.175 | ndrg2 | -0.057 |
| s100a1 | 0.175 | rpl30 | -0.056 |
| frem1b | 0.172 | rps21 | -0.055 |