ubiquitin-conjugating enzyme E2T (putative)
ZFIN 
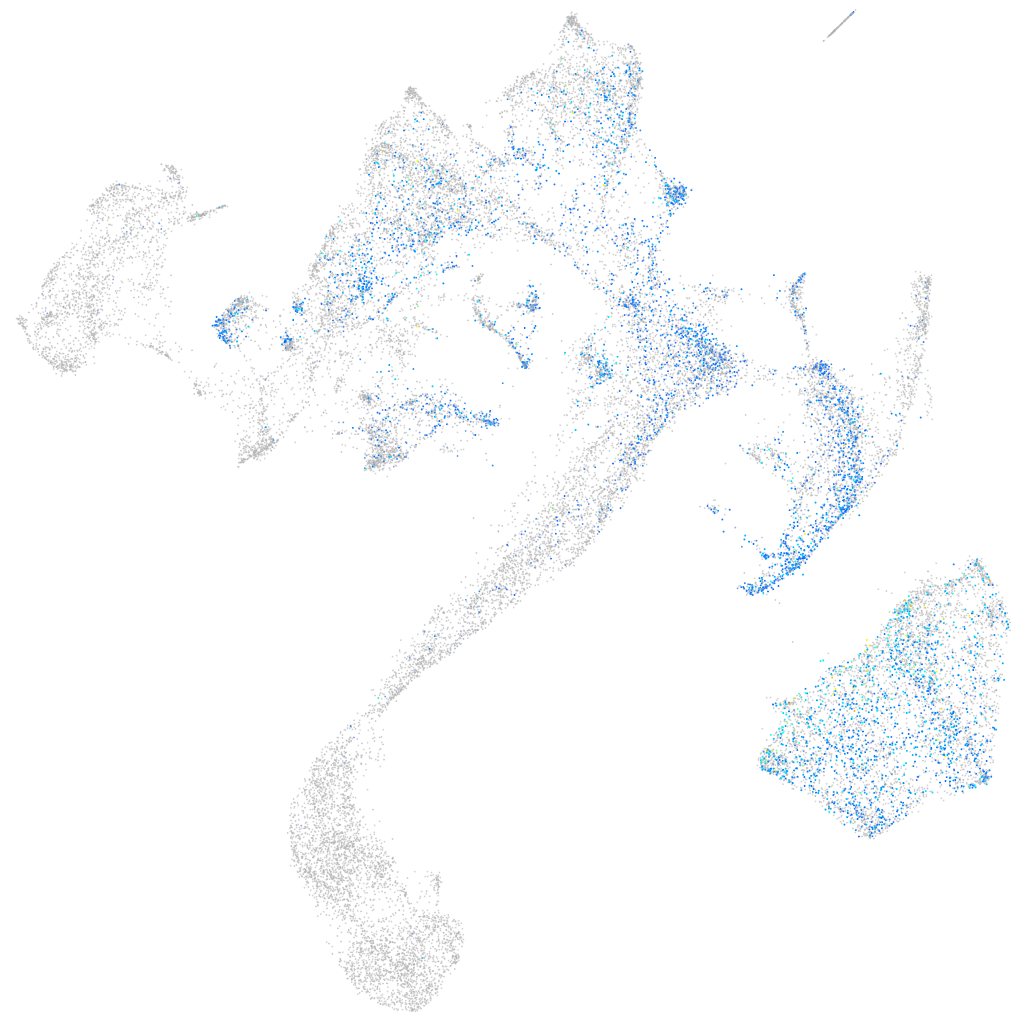
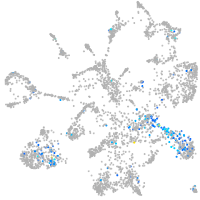
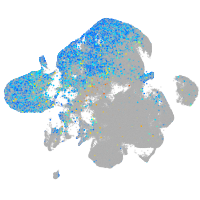
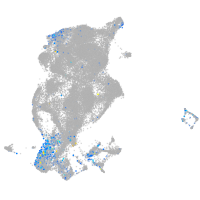
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pcna | 0.384 | actc1b | -0.262 |
| chaf1a | 0.340 | ttn.2 | -0.238 |
| stmn1a | 0.340 | aldoab | -0.236 |
| tubb2b | 0.339 | ttn.1 | -0.236 |
| tuba8l4 | 0.335 | ak1 | -0.232 |
| nasp | 0.333 | atp2a1 | -0.230 |
| rbbp4 | 0.333 | tmem38a | -0.229 |
| rrm1 | 0.333 | ckmb | -0.227 |
| anp32b | 0.331 | ckma | -0.227 |
| fen1 | 0.331 | tnnc2 | -0.223 |
| rpa2 | 0.327 | acta1b | -0.218 |
| hmgb2a | 0.326 | neb | -0.215 |
| sumo3b | 0.324 | desma | -0.214 |
| seta | 0.322 | mybphb | -0.213 |
| zgc:110540 | 0.322 | srl | -0.213 |
| rpa3 | 0.320 | acta1a | -0.212 |
| dek | 0.319 | tpma | -0.212 |
| banf1 | 0.318 | mylpfa | -0.211 |
| si:ch211-222l21.1 | 0.317 | ldb3a | -0.211 |
| dhfr | 0.314 | pabpc4 | -0.210 |
| lig1 | 0.314 | gapdh | -0.208 |
| hmgb2b | 0.311 | CABZ01078594.1 | -0.207 |
| slbp | 0.308 | eno1a | -0.204 |
| cbx3a | 0.302 | cav3 | -0.204 |
| mibp | 0.300 | ldb3b | -0.204 |
| hmga1a | 0.299 | vdac3 | -0.201 |
| mcm7 | 0.297 | nme2b.2 | -0.201 |
| cirbpa | 0.295 | actn3b | -0.200 |
| ranbp1 | 0.295 | gamt | -0.199 |
| hnrnpabb | 0.292 | eno3 | -0.196 |
| ssrp1a | 0.290 | si:ch73-367p23.2 | -0.196 |
| ptmab | 0.290 | myl1 | -0.195 |
| esco2 | 0.289 | mylz3 | -0.194 |
| sumo3a | 0.288 | actn3a | -0.194 |
| CABZ01005379.1 | 0.287 | txlnbb | -0.194 |