ubiquitin-like modifier activating enzyme 1
ZFIN 
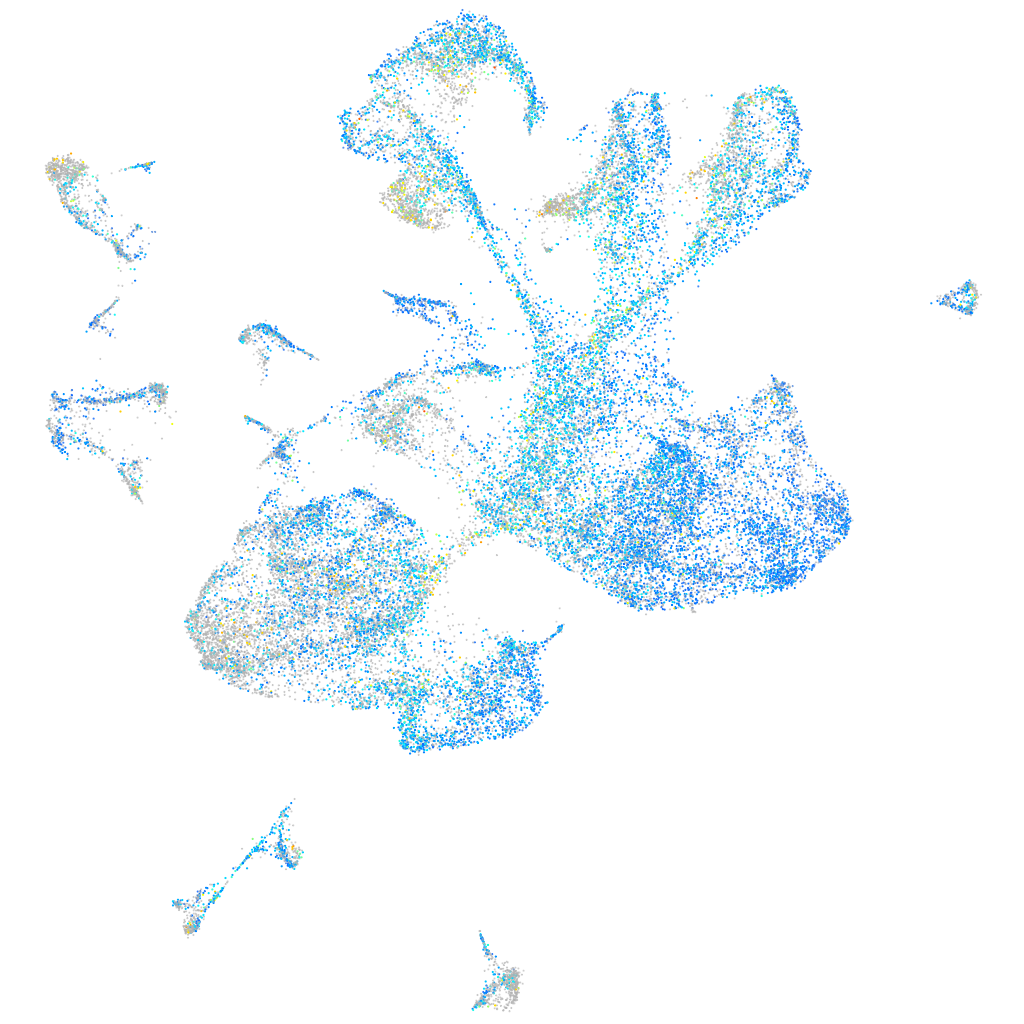
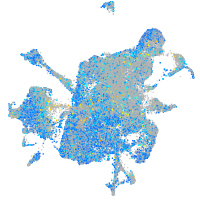
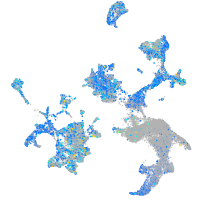
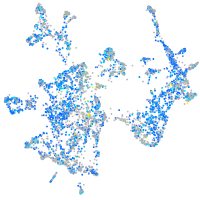
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hnrnpaba | 0.219 | glula | -0.146 |
| khdrbs1a | 0.212 | atp1a1b | -0.144 |
| hnrnpa0l | 0.204 | slc1a2b | -0.140 |
| ptmab | 0.197 | ptn | -0.133 |
| cirbpa | 0.196 | efhd1 | -0.132 |
| hnrnpa0b | 0.194 | cx43 | -0.131 |
| tubb2b | 0.193 | si:ch211-66e2.5 | -0.130 |
| si:ch211-222l21.1 | 0.193 | ppap2d | -0.127 |
| cirbpb | 0.192 | fabp7a | -0.124 |
| hdac1 | 0.191 | slc4a4a | -0.121 |
| hnrnpabb | 0.184 | slc3a2a | -0.119 |
| tubb5 | 0.183 | cebpd | -0.118 |
| si:ch211-288g17.3 | 0.182 | cox4i2 | -0.118 |
| ddx39ab | 0.177 | mdkb | -0.117 |
| hmga1a | 0.177 | gpr37l1b | -0.116 |
| ilf2 | 0.176 | mt2 | -0.116 |
| marcksb | 0.176 | hepacama | -0.114 |
| hspa8 | 0.173 | dap1b | -0.114 |
| hnrnpa0a | 0.173 | cdo1 | -0.108 |
| seta | 0.170 | apoa2 | -0.107 |
| h3f3d | 0.167 | ptgdsb.2 | -0.105 |
| nono | 0.166 | acbd7 | -0.105 |
| syncrip | 0.166 | slc6a1b | -0.101 |
| hnrnpub | 0.165 | cd63 | -0.100 |
| smarce1 | 0.165 | qki2 | -0.100 |
| snrpd2 | 0.165 | sept8b | -0.100 |
| smarca4a | 0.164 | cyp2ad3 | -0.100 |
| sox11b | 0.163 | mfge8a | -0.100 |
| sumo3a | 0.163 | slc1a3b | -0.098 |
| nucks1a | 0.163 | anxa13 | -0.097 |
| h2afvb | 0.161 | cspg5b | -0.097 |
| hnrnph1l | 0.161 | apoa1b | -0.096 |
| h3f3a | 0.159 | nck1a | -0.096 |
| hmgb2b | 0.159 | zgc:153704 | -0.095 |
| alyref | 0.159 | eno1b | -0.095 |