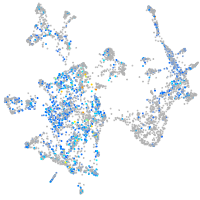
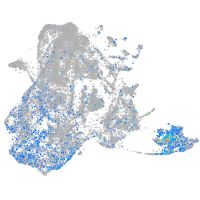
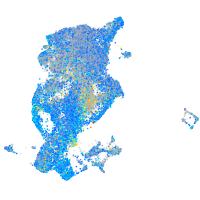
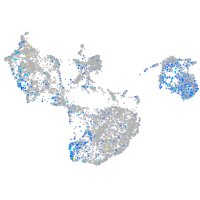
UDP-N-acetylglucosamine pyrophosphorylase 1
ZFIN 
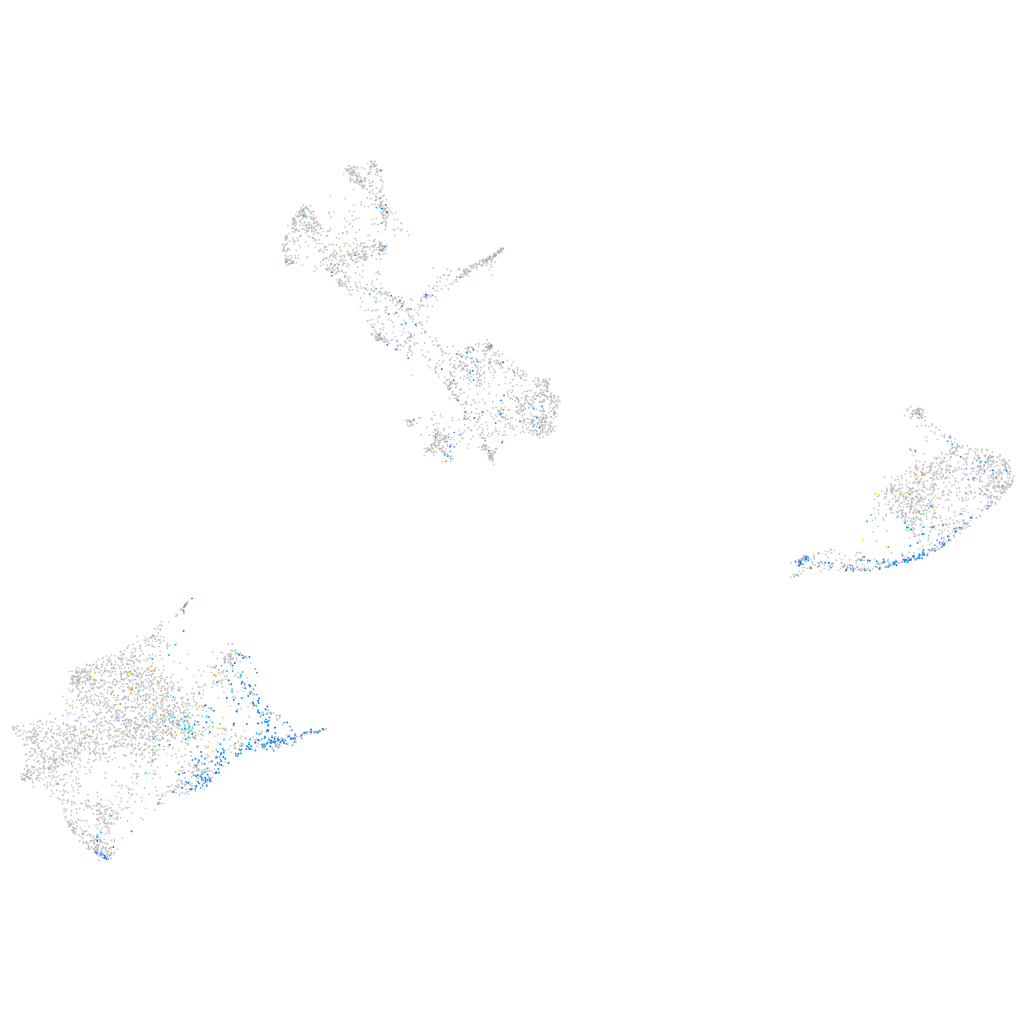
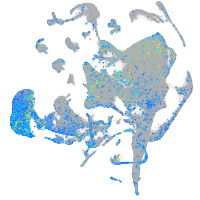
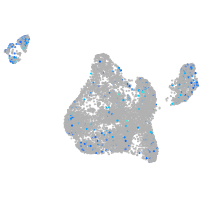
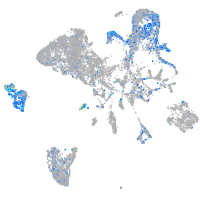
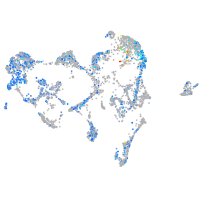
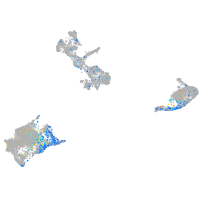
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sprb | 0.186 | pvalb2 | -0.107 |
| cx30.3 | 0.184 | pvalb1 | -0.104 |
| rgs2 | 0.173 | apoda.1 | -0.089 |
| ssr4 | 0.168 | gpnmb | -0.088 |
| xbp1 | 0.167 | defbl1 | -0.087 |
| XLOC-001964 | 0.164 | si:ch211-256m1.8 | -0.083 |
| pts | 0.157 | lypc | -0.081 |
| prdx1 | 0.155 | mdkb | -0.076 |
| lman2 | 0.154 | si:dkey-197i20.6 | -0.075 |
| akr1b1 | 0.152 | LOC103909099 | -0.075 |
| pcbd1 | 0.151 | akap12a | -0.074 |
| gpd1b | 0.148 | unm-sa821 | -0.074 |
| mitfa | 0.148 | cdh11 | -0.073 |
| mocs1 | 0.145 | alx4a | -0.073 |
| qdpra | 0.144 | cox4i2 | -0.073 |
| cst14b.1 | 0.144 | actc1b | -0.073 |
| C12orf75 | 0.144 | si:ch73-89b15.3 | -0.072 |
| pah | 0.144 | apoa1b | -0.072 |
| phlda1 | 0.144 | dap1b | -0.072 |
| tmem243a | 0.141 | mylz3 | -0.072 |
| canx | 0.141 | sdc2 | -0.070 |
| tram1 | 0.140 | alx4b | -0.069 |
| pax7a | 0.139 | myhz1.1 | -0.068 |
| crestin | 0.138 | ptmaa | -0.067 |
| ssr2 | 0.137 | prx | -0.067 |
| rbp4l | 0.137 | s100v2 | -0.067 |
| sigmar1 | 0.135 | ak1 | -0.066 |
| si:dkeyp-110e4.6 | 0.135 | apoa2 | -0.065 |
| CABZ01032488.1 | 0.134 | prss59.2 | -0.065 |
| cax1 | 0.134 | sytl2b | -0.065 |
| naga | 0.134 | tuba1c | -0.064 |
| atp6v1ab | 0.134 | ponzr1 | -0.063 |
| rab5if | 0.134 | alx1 | -0.063 |
| itga4 | 0.133 | slc29a1a | -0.062 |
| tspan36 | 0.133 | ndrg2 | -0.062 |