tRNA-yW synthesizing protein 3 homolog (S. cerevisiae)
ZFIN 
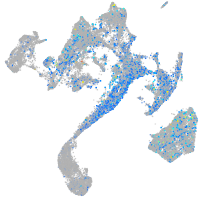
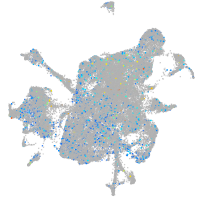
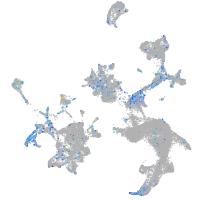
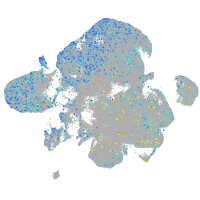
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CABZ01057381.1 | 0.150 | col1a1a | -0.072 |
| syt17 | 0.146 | tmsb4x | -0.056 |
| BX000363.1 | 0.140 | and1 | -0.055 |
| CU855685.1 | 0.123 | si:ch211-246m6.5 | -0.048 |
| npm1a | 0.120 | and2 | -0.048 |
| slc47a4 | 0.119 | ost4 | -0.047 |
| bysl | 0.118 | actb1 | -0.046 |
| CABZ01069436.1 | 0.117 | fkbp9 | -0.046 |
| LOC101884285 | 0.112 | frem2a | -0.045 |
| gtpbp4 | 0.112 | cap1 | -0.045 |
| BX119909.1 | 0.112 | CABZ01074130.1 | -0.044 |
| zgc:109982 | 0.111 | vcanb | -0.044 |
| CABZ01016118.1 | 0.111 | rap1b | -0.043 |
| rsl1d1 | 0.108 | sec24d | -0.042 |
| AL954696.2 | 0.108 | vcana | -0.042 |
| tspan10 | 0.105 | XLOC-013086 | -0.041 |
| CABZ01049825.1 | 0.105 | fstl1a | -0.041 |
| eif3ba | 0.102 | bcam | -0.039 |
| XLOC-018363 | 0.101 | actn1 | -0.039 |
| BX890539.1 | 0.101 | mdka | -0.039 |
| eif4ebp3l | 0.101 | efhd2 | -0.038 |
| tcp1 | 0.101 | myoc | -0.038 |
| CU655820.1 | 0.101 | prdx1 | -0.038 |
| PITPNM2 | 0.101 | tax1bp3 | -0.038 |
| btf3 | 0.098 | gsnb | -0.037 |
| cct7 | 0.098 | pls3 | -0.037 |
| CABZ01064771.1 | 0.097 | alox5a | -0.037 |
| naca | 0.097 | hbae1.1 | -0.037 |
| si:ch211-217k17.7 | 0.096 | myh9a | -0.037 |
| cct3 | 0.096 | ext1a | -0.036 |
| gdf5 | 0.095 | hbbe1.3 | -0.036 |
| rsl24d1 | 0.094 | si:ch211-243g18.2 | -0.036 |
| cct5 | 0.094 | thy1 | -0.036 |
| npm3 | 0.094 | pah | -0.036 |
| robo4 | 0.094 | rhbdf1a | -0.035 |