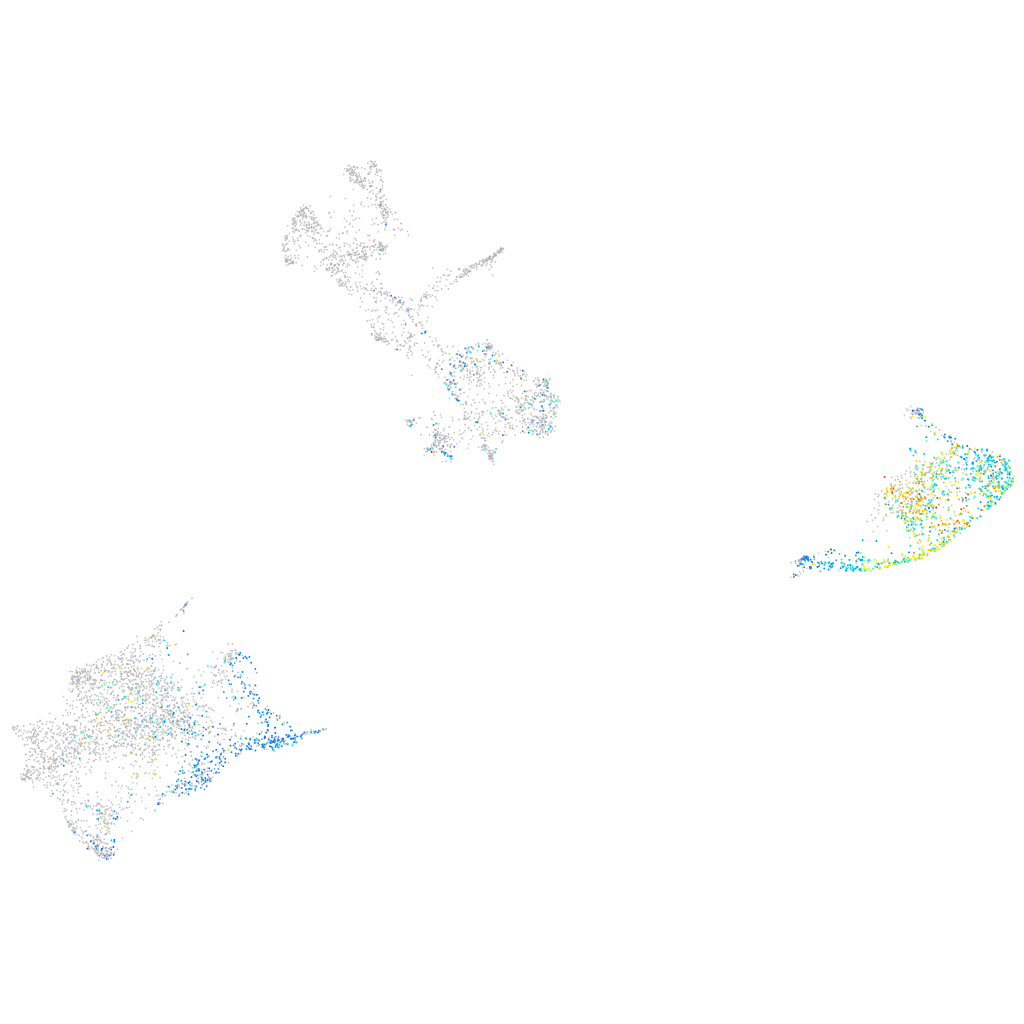
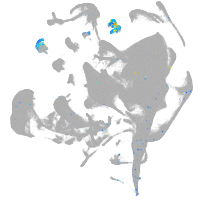
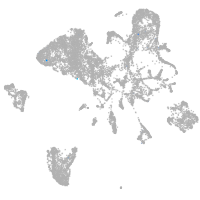
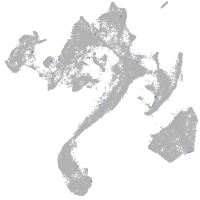
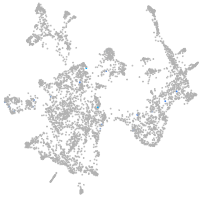
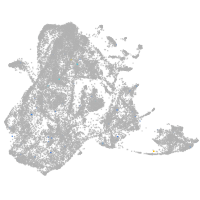
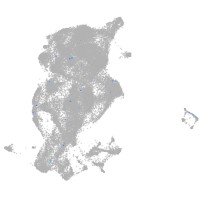
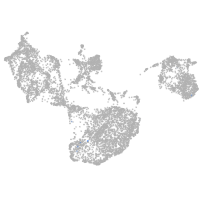
tyrosinase
ZFIN 
Other cell groups


Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pmela | 0.705 | paics | -0.332 |
| tyrp1a | 0.702 | CABZ01021592.1 | -0.317 |
| dct | 0.699 | uraha | -0.304 |
| tyrp1b | 0.695 | phyhd1 | -0.278 |
| oca2 | 0.666 | mdh1aa | -0.274 |
| zgc:91968 | 0.645 | si:dkey-251i10.2 | -0.267 |
| slc24a5 | 0.599 | defbl1 | -0.244 |
| mtbl | 0.585 | apoda.1 | -0.241 |
| kita | 0.566 | krt18b | -0.224 |
| slc45a2 | 0.560 | gpnmb | -0.220 |
| si:ch73-389b16.1 | 0.546 | si:ch211-256m1.8 | -0.217 |
| prkar1b | 0.542 | tmem130 | -0.214 |
| aadac | 0.520 | lypc | -0.213 |
| gstt1a | 0.516 | si:dkey-197i20.6 | -0.210 |
| tspan36 | 0.512 | unm-sa821 | -0.204 |
| lamp1a | 0.503 | slc2a15a | -0.203 |
| tspan10 | 0.502 | mdkb | -0.201 |
| pah | 0.484 | impdh1b | -0.197 |
| atp6v0ca | 0.483 | atic | -0.197 |
| slc37a2 | 0.482 | glulb | -0.194 |
| slc29a3 | 0.478 | alx4a | -0.193 |
| SPAG9 | 0.471 | sparc | -0.189 |
| slc7a5 | 0.471 | si:ch73-89b15.3 | -0.189 |
| kcnj13 | 0.470 | si:ch211-251b21.1 | -0.189 |
| slc22a2 | 0.470 | akap12a | -0.187 |
| anxa1a | 0.465 | mdka | -0.187 |
| slc3a2a | 0.464 | prps1a | -0.185 |
| slc39a10 | 0.461 | s100v2 | -0.180 |
| qdpra | 0.447 | alx4b | -0.179 |
| rab38 | 0.440 | ponzr1 | -0.175 |
| rabl6b | 0.434 | pltp | -0.172 |
| slc24a4a | 0.431 | cdh11 | -0.170 |
| tfap2e | 0.429 | sytl2b | -0.169 |
| mitfa | 0.420 | tcirg1a | -0.169 |
| tmem243b | 0.416 | prdx5 | -0.167 |