"tubulin, alpha 8 like 2"
ZFIN 
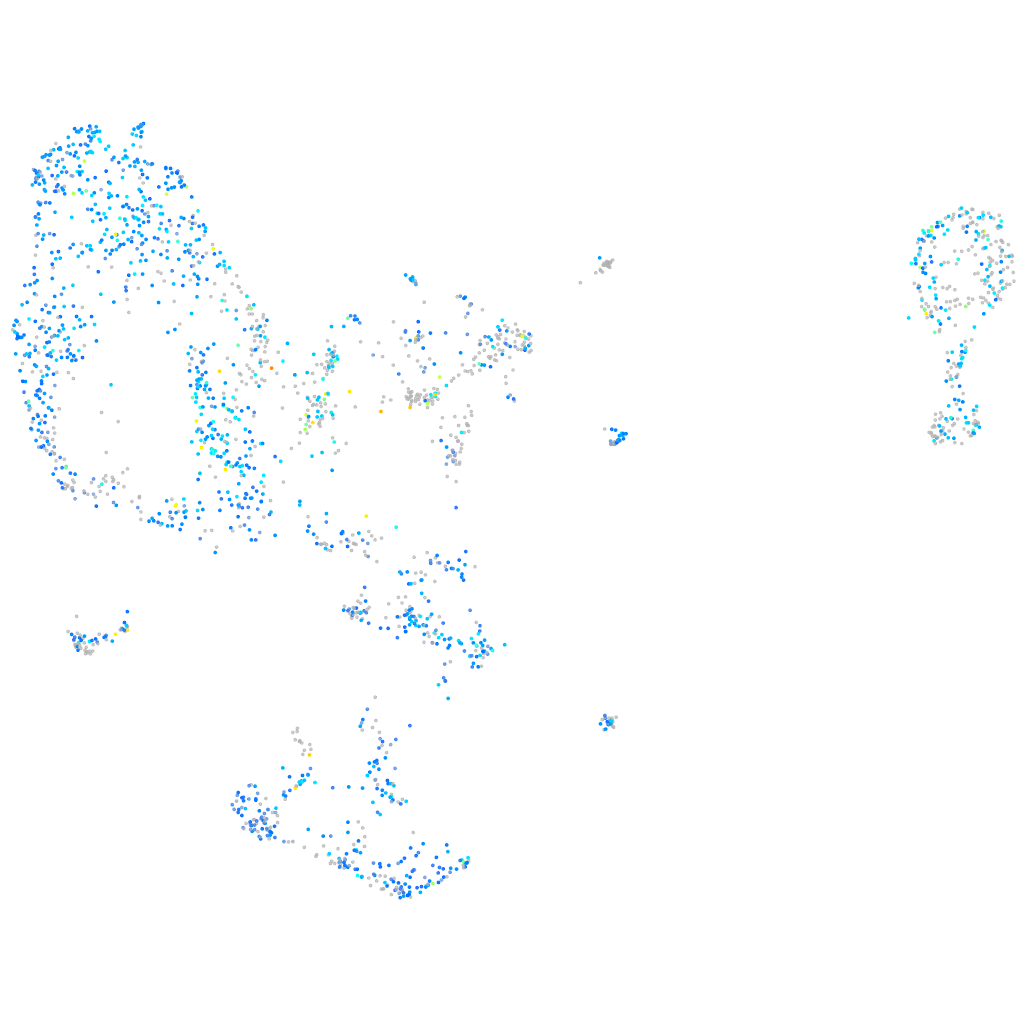
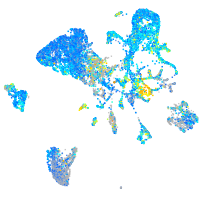
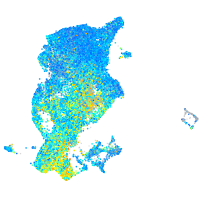
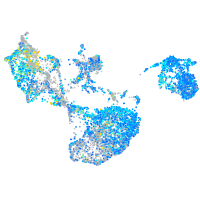
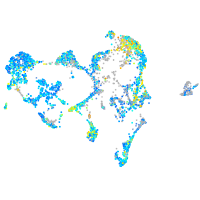
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gpd1b | 0.279 | marcksl1b | -0.162 |
| zgc:136493 | 0.277 | si:ch73-1a9.3 | -0.148 |
| suclg2 | 0.274 | mdkb | -0.147 |
| si:dkey-28n18.9 | 0.274 | hnrnpa0a | -0.140 |
| suclg1 | 0.271 | nova2 | -0.140 |
| nrip2 | 0.270 | gpm6ab | -0.138 |
| rnaseka | 0.268 | syt1a | -0.138 |
| dab2 | 0.265 | rtn1a | -0.137 |
| si:dkey-103j14.5 | 0.260 | tuba1c | -0.137 |
| ctsla | 0.260 | jpt1b | -0.135 |
| slc6a19a.1 | 0.256 | hmgb1b | -0.134 |
| zgc:101040 | 0.255 | celf2 | -0.128 |
| glyctk | 0.255 | elavl3 | -0.128 |
| pklr | 0.254 | dbn1 | -0.126 |
| gpt2l | 0.253 | dpysl2b | -0.126 |
| grhprb | 0.252 | gapdhs | -0.125 |
| gcshb | 0.251 | jam3b | -0.124 |
| upb1 | 0.251 | marcksl1a | -0.123 |
| slc37a4a | 0.250 | CU467822.1 | -0.121 |
| ctsz | 0.249 | tmsb | -0.119 |
| sord | 0.248 | sfrp1a | -0.119 |
| CU682777.2 | 0.248 | hmgb3a | -0.116 |
| gstp1 | 0.248 | stmn1b | -0.115 |
| nit2 | 0.247 | gpm6aa | -0.115 |
| pdzk1 | 0.246 | vamp2 | -0.114 |
| scpep1 | 0.245 | hmgb1a | -0.113 |
| acy3.2 | 0.244 | ap1s2 | -0.113 |
| akr7a3 | 0.243 | ppm1la | -0.112 |
| si:dkey-194e6.1 | 0.242 | sncb | -0.111 |
| aldh7a1 | 0.242 | tmeff1b | -0.111 |
| cubn | 0.241 | hmgn2 | -0.110 |
| epdl2 | 0.241 | ckbb | -0.110 |
| tmprss2 | 0.241 | si:ch211-137a8.4 | -0.108 |
| eno3 | 0.240 | nanos1 | -0.108 |
| slc2a2 | 0.240 | tusc3 | -0.107 |