"tubulin, alpha 1c"
ZFIN 
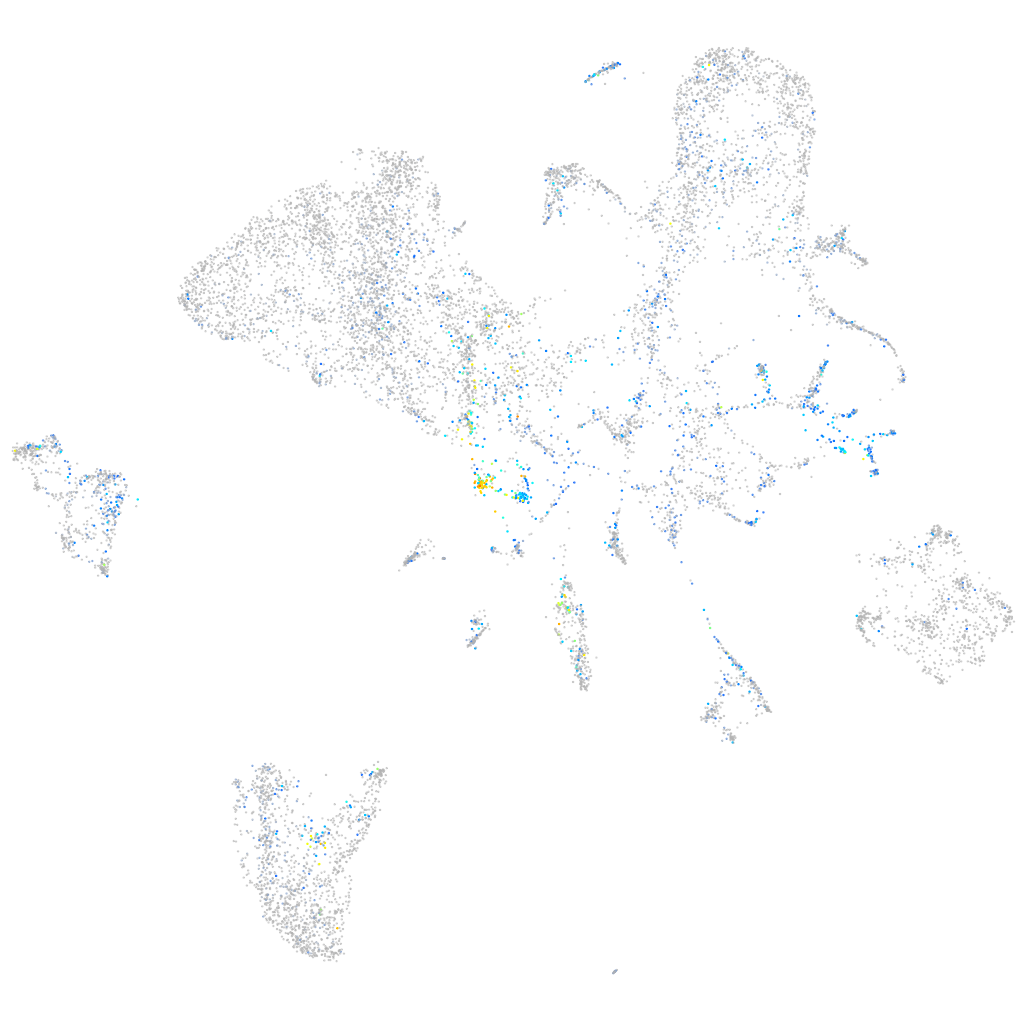
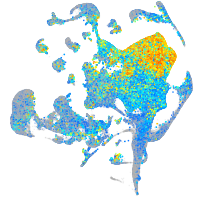
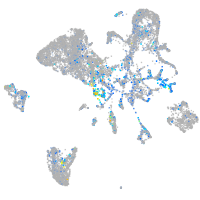
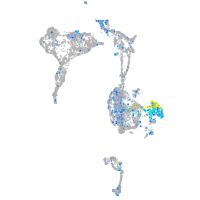
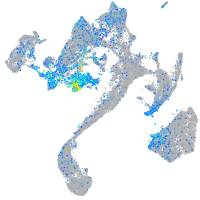
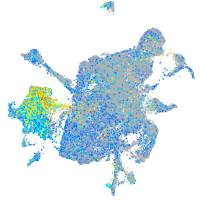
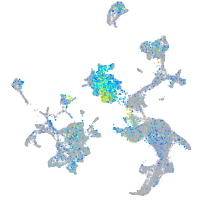
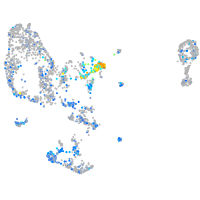
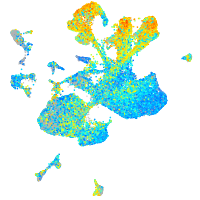
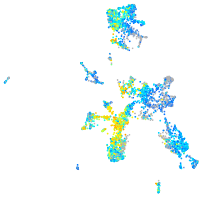
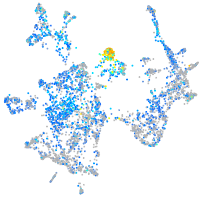
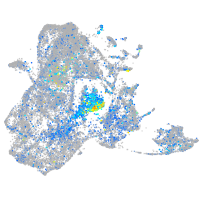
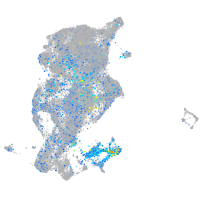
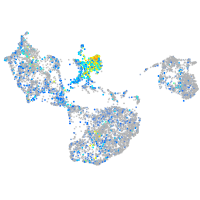
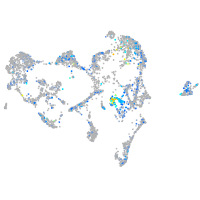
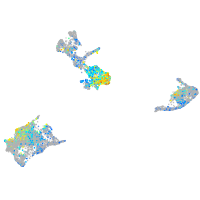
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| elavl3 | 0.447 | aldob | -0.170 |
| stmn1b | 0.434 | aldh6a1 | -0.166 |
| gng3 | 0.400 | glud1b | -0.165 |
| nova2 | 0.389 | cx32.3 | -0.161 |
| gpm6aa | 0.389 | pnp4b | -0.159 |
| tuba1a | 0.388 | nupr1b | -0.154 |
| sncb | 0.377 | gapdh | -0.153 |
| rtn1b | 0.369 | abat | -0.151 |
| stxbp1a | 0.365 | hdlbpa | -0.150 |
| elavl4 | 0.361 | fbp1b | -0.146 |
| tubb5 | 0.349 | gamt | -0.146 |
| cspg5a | 0.349 | scp2a | -0.145 |
| fez1 | 0.325 | g6pca.2 | -0.144 |
| tmsb | 0.324 | rdh1 | -0.142 |
| gnao1a | 0.324 | cyp4v8 | -0.142 |
| mdkb | 0.323 | dhrs9 | -0.140 |
| gap43 | 0.312 | cox7a1 | -0.139 |
| stmn2a | 0.311 | gstr | -0.138 |
| celf2 | 0.310 | agxtb | -0.138 |
| cnrip1a | 0.308 | sdr16c5b | -0.137 |
| dpysl3 | 0.305 | mat1a | -0.136 |
| myt1b | 0.303 | mgst1.2 | -0.135 |
| rnasekb | 0.302 | gatm | -0.134 |
| CU467822.1 | 0.302 | ckba | -0.134 |
| dpysl2b | 0.301 | tdo2a | -0.134 |
| ptmaa | 0.295 | cpn1 | -0.133 |
| csdc2a | 0.290 | eno3 | -0.132 |
| ywhag2 | 0.287 | haao | -0.132 |
| stx1b | 0.285 | eef1da | -0.132 |
| snap25a | 0.284 | nipsnap3a | -0.132 |
| cadm3 | 0.283 | etnppl | -0.132 |
| vamp2 | 0.282 | cebpd | -0.132 |
| celf4 | 0.282 | agxta | -0.131 |
| zgc:65894 | 0.281 | aldh1l1 | -0.131 |
| LOC100537384 | 0.281 | aldh7a1 | -0.131 |