"tubulin tyrosine ligase-like family, member 7"
ZFIN 
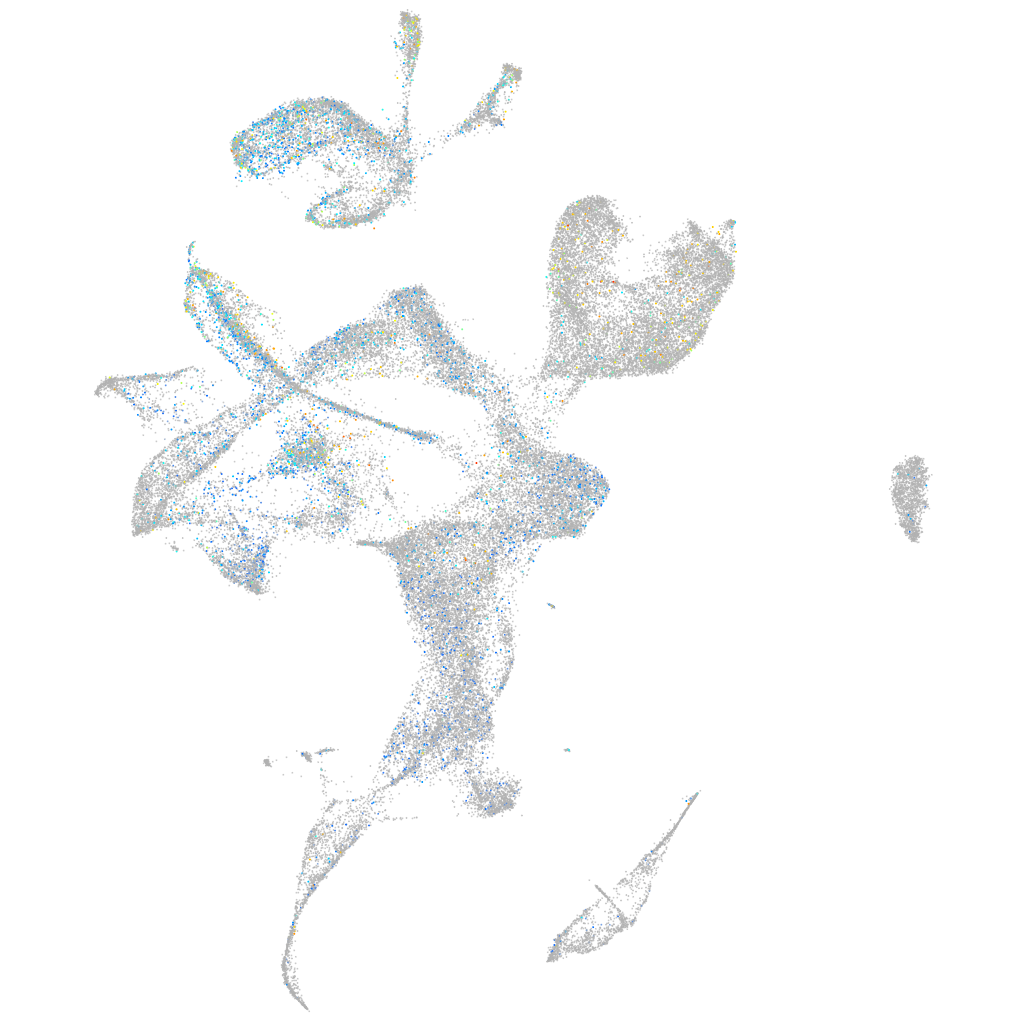
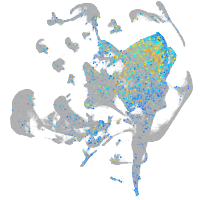
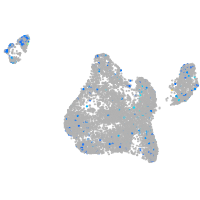
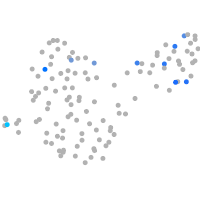
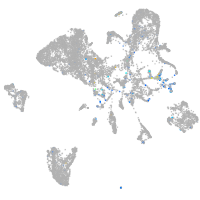
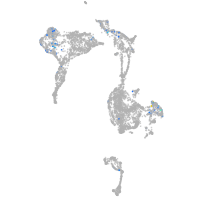
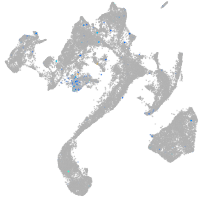
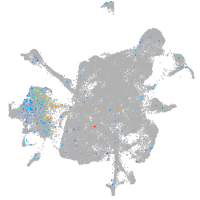
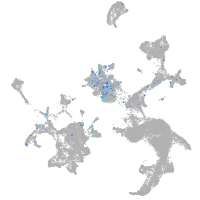
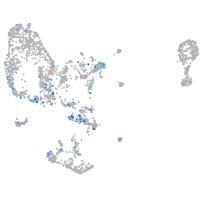
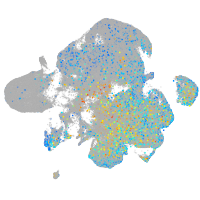
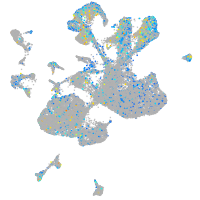
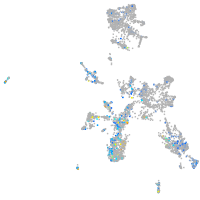
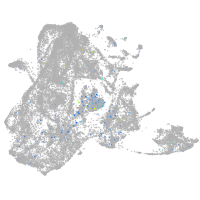
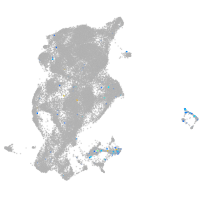
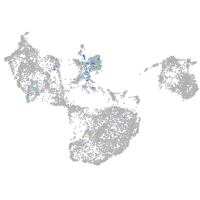
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| elavl3 | 0.142 | hmgb2a | -0.100 |
| stxbp1a | 0.140 | pcna | -0.069 |
| stx1b | 0.139 | id1 | -0.066 |
| syt1a | 0.136 | mcm7 | -0.065 |
| sv2a | 0.131 | selenoh | -0.065 |
| eno2 | 0.130 | rrm1 | -0.065 |
| ywhag2 | 0.130 | fen1 | -0.063 |
| elavl4 | 0.126 | chaf1a | -0.062 |
| rtn1b | 0.126 | ahcy | -0.062 |
| sncb | 0.125 | mcm6 | -0.062 |
| stmn2a | 0.121 | cdca7b | -0.061 |
| zgc:65894 | 0.121 | nutf2l | -0.061 |
| gnao1a | 0.118 | nasp | -0.061 |
| rbfox1 | 0.114 | mcm2 | -0.060 |
| stmn1b | 0.114 | mcm5 | -0.059 |
| syt2a | 0.114 | mcm3 | -0.058 |
| gng3 | 0.113 | dkc1 | -0.058 |
| gabrb4 | 0.113 | lig1 | -0.057 |
| tuba2 | 0.109 | cdca7a | -0.057 |
| myt1la | 0.109 | COX7A2 (1 of many) | -0.057 |
| carmil2 | 0.107 | mdka | -0.057 |
| mir181b-3 | 0.106 | tuba8l4 | -0.057 |
| zgc:153426 | 0.106 | banf1 | -0.057 |
| si:ch211-129p13.1 | 0.105 | stmn1a | -0.056 |
| basp1 | 0.104 | msna | -0.056 |
| snap25a | 0.104 | rpa2 | -0.056 |
| zfhx3 | 0.104 | anp32b | -0.056 |
| si:ch73-119p20.1 | 0.103 | mcm4 | -0.056 |
| slc32a1 | 0.103 | dut | -0.055 |
| nrxn2a | 0.102 | ccnd1 | -0.055 |
| sprn | 0.102 | CABZ01005379.1 | -0.055 |
| id4 | 0.101 | hmgb2b | -0.055 |
| vamp2 | 0.101 | cx43.4 | -0.054 |
| cplx2 | 0.100 | slbp | -0.054 |
| tmsb2 | 0.100 | ranbp1 | -0.054 |