"transient receptor potential cation channel, subfamily C, member 7b"
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm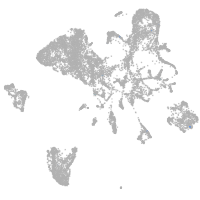 |
axial mesoderm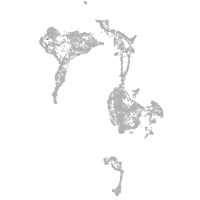 |
muscle  |
mural cells / non-skeletal muscle 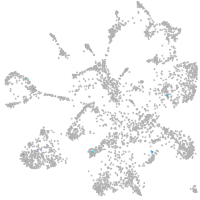 |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin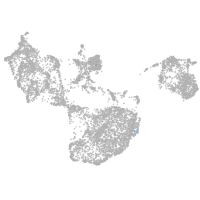 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg2b | 0.085 | aldob | -0.025 |
| ywhag2 | 0.085 | hspb1 | -0.021 |
| zgc:65894 | 0.082 | apoc1 | -0.020 |
| sncb | 0.080 | apoeb | -0.020 |
| stmn2a | 0.079 | lye | -0.019 |
| sv2a | 0.078 | si:ch211-152c2.3 | -0.019 |
| snap25a | 0.074 | stm | -0.019 |
| stx1b | 0.074 | wu:fb18f06 | -0.019 |
| vamp2 | 0.073 | akap12b | -0.018 |
| slc6a1a | 0.072 | anxa1c | -0.018 |
| elavl4 | 0.071 | arf1 | -0.018 |
| map1aa | 0.071 | cast | -0.018 |
| rtn1b | 0.070 | cx43.4 | -0.018 |
| syn2a | 0.070 | hdlbpa | -0.018 |
| stxbp1a | 0.070 | ppl | -0.018 |
| atp6v0cb | 0.069 | tspo | -0.018 |
| atp6v1b2 | 0.069 | zgc:193505 | -0.018 |
| cdk5r2a | 0.068 | capn9 | -0.017 |
| eno2 | 0.067 | ccng1 | -0.017 |
| gap43 | 0.067 | cst14b.1 | -0.017 |
| syngr3a | 0.067 | evpla | -0.017 |
| dnajc5aa | 0.066 | icn2 | -0.017 |
| gng3 | 0.066 | mgst1.2 | -0.017 |
| nova1 | 0.066 | nop58 | -0.017 |
| vsnl1b | 0.066 | npm1a | -0.017 |
| camk2n1a | 0.065 | polr3gla | -0.017 |
| gnao1a | 0.065 | si:ch211-125o16.4 | -0.017 |
| mllt11 | 0.065 | tuba8l2 | -0.017 |
| nrxn1a | 0.065 | znf185 | -0.017 |
| pcsk1nl | 0.065 | agr1 | -0.016 |
| si:ch73-119p20.1 | 0.065 | anxa1b | -0.016 |
| tuba2 | 0.065 | cldne | -0.016 |
| cplx2 | 0.064 | dkc1 | -0.016 |
| gabrb4 | 0.064 | elovl7a | -0.016 |
| atp2b3a | 0.063 | fbl | -0.016 |




