tumor protein p63 regulated 1-like
ZFIN 
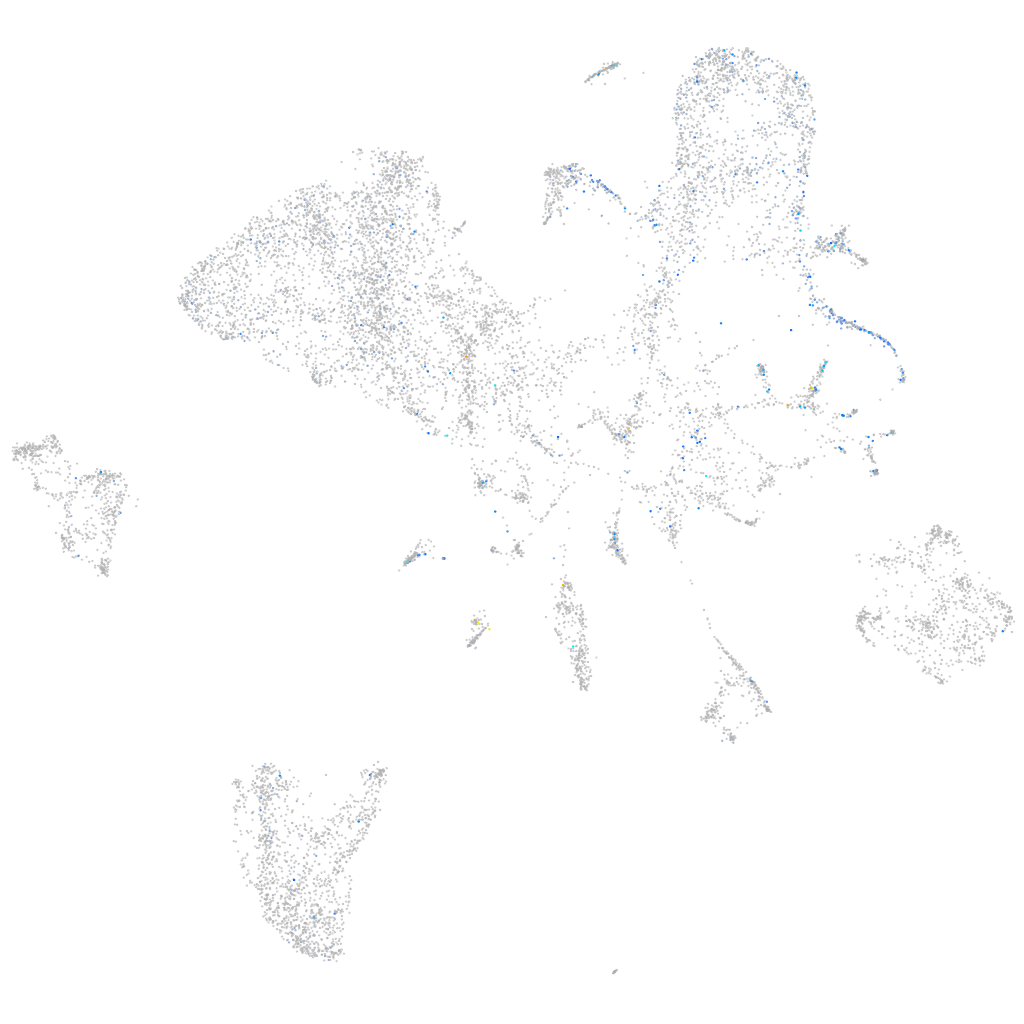
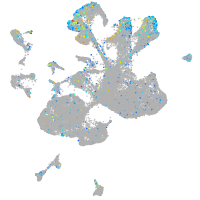
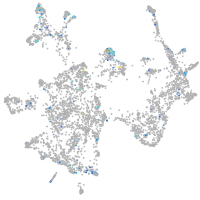
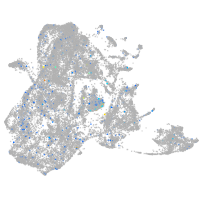
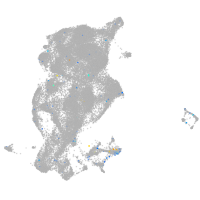
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tmem176l.2 | 0.171 | apoc1 | -0.080 |
| si:ch211-137i24.10 | 0.166 | bhmt | -0.078 |
| wu:fb59d01 | 0.163 | fabp3 | -0.068 |
| tmem45b | 0.161 | aqp12 | -0.068 |
| eps8l3b | 0.154 | agxtb | -0.065 |
| abca2 | 0.151 | ttc36 | -0.059 |
| RF00478 | 0.147 | mat1a | -0.059 |
| calml4a | 0.146 | tfa | -0.058 |
| lgals2b | 0.145 | apoa2 | -0.057 |
| cldn15la | 0.142 | cpn1 | -0.057 |
| si:dkeyp-73b11.8 | 0.142 | grhprb | -0.056 |
| c1qa | 0.140 | hspb1 | -0.056 |
| prr15la | 0.139 | serpina1 | -0.056 |
| scg3 | 0.138 | gamt | -0.056 |
| itpk1a | 0.136 | fetub | -0.056 |
| gucy2c | 0.134 | ambp | -0.056 |
| tmem59 | 0.134 | fabp10a | -0.055 |
| si:dkeyp-72e1.6 | 0.133 | apom | -0.055 |
| pllp | 0.133 | serpina1l | -0.055 |
| vil1 | 0.130 | zgc:123103 | -0.055 |
| psmb11b | 0.130 | fgb | -0.055 |
| trim35-12 | 0.128 | gatm | -0.054 |
| LOC100534786 | 0.128 | hao1 | -0.054 |
| si:dkey-283b1.6 | 0.128 | ces2 | -0.054 |
| st3gal7 | 0.128 | LOC110437731 | -0.054 |
| lrrfip1b | 0.127 | ttr | -0.054 |
| zgc:136872 | 0.127 | ndrg2 | -0.054 |
| si:ch1073-443f11.2 | 0.127 | rbp2b | -0.053 |
| myo15b | 0.126 | rbp4 | -0.053 |
| degs2 | 0.125 | kng1 | -0.053 |
| igsf9a | 0.125 | fga | -0.053 |
| atp10d | 0.124 | akap12b | -0.053 |
| zgc:172079 | 0.122 | fgg | -0.053 |
| zgc:165573 | 0.122 | npm1a | -0.052 |
| tm4sf4 | 0.122 | vtnb | -0.052 |