tumor protein p53 inducible nuclear protein 1
ZFIN 
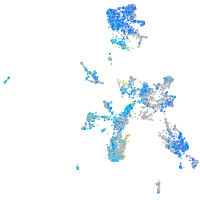
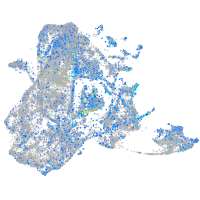
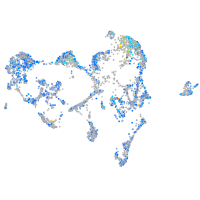
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CR383676.1 | 0.167 | cox7c | -0.114 |
| epcam | 0.161 | tma7 | -0.112 |
| btg1 | 0.155 | rpl38 | -0.104 |
| tmem59 | 0.154 | foxi3a | -0.097 |
| spint2 | 0.147 | gcm2 | -0.096 |
| ier5 | 0.145 | npm1a | -0.094 |
| cd63 | 0.140 | atp5mf | -0.092 |
| BX000438.2 | 0.140 | atp5f1e | -0.090 |
| si:dkey-193p11.2 | 0.139 | mt-atp8 | -0.090 |
| litaf | 0.138 | rpia | -0.089 |
| gabarapb | 0.137 | rpl36a | -0.089 |
| zgc:100829 | 0.137 | nop58 | -0.089 |
| malb | 0.136 | atp6v1ba | -0.088 |
| chmp1b | 0.133 | rpl37 | -0.087 |
| krt91 | 0.130 | rab5if | -0.086 |
| si:ch211-260e23.9 | 0.129 | rps28 | -0.085 |
| tuft1a | 0.129 | atp5md | -0.084 |
| cldnb | 0.129 | rasgrp4 | -0.083 |
| aldob | 0.128 | mrpl12 | -0.083 |
| pawr | 0.127 | polr2k | -0.083 |
| jun | 0.127 | cox5ab | -0.083 |
| ier2b | 0.126 | etf1a | -0.082 |
| ccng2 | 0.125 | cox8a | -0.082 |
| ubb | 0.124 | vars | -0.080 |
| selenow1 | 0.124 | cox6c | -0.080 |
| map1lc3a | 0.124 | desi2 | -0.079 |
| nupr1b | 0.123 | cox7a2a | -0.079 |
| mcl1a | 0.122 | imp4 | -0.077 |
| ucp2 | 0.121 | ca15a | -0.077 |
| fa2h | 0.120 | atp5if1a | -0.077 |
| myzap | 0.119 | slc6a6b | -0.077 |
| eif1b | 0.119 | atp6v0a1a | -0.077 |
| btg2 | 0.118 | zgc:193541 | -0.077 |
| junba | 0.117 | uqcrb | -0.076 |
| tjp2b | 0.117 | snrpd1 | -0.076 |