tumor protein p53 inducible protein 11a
ZFIN 
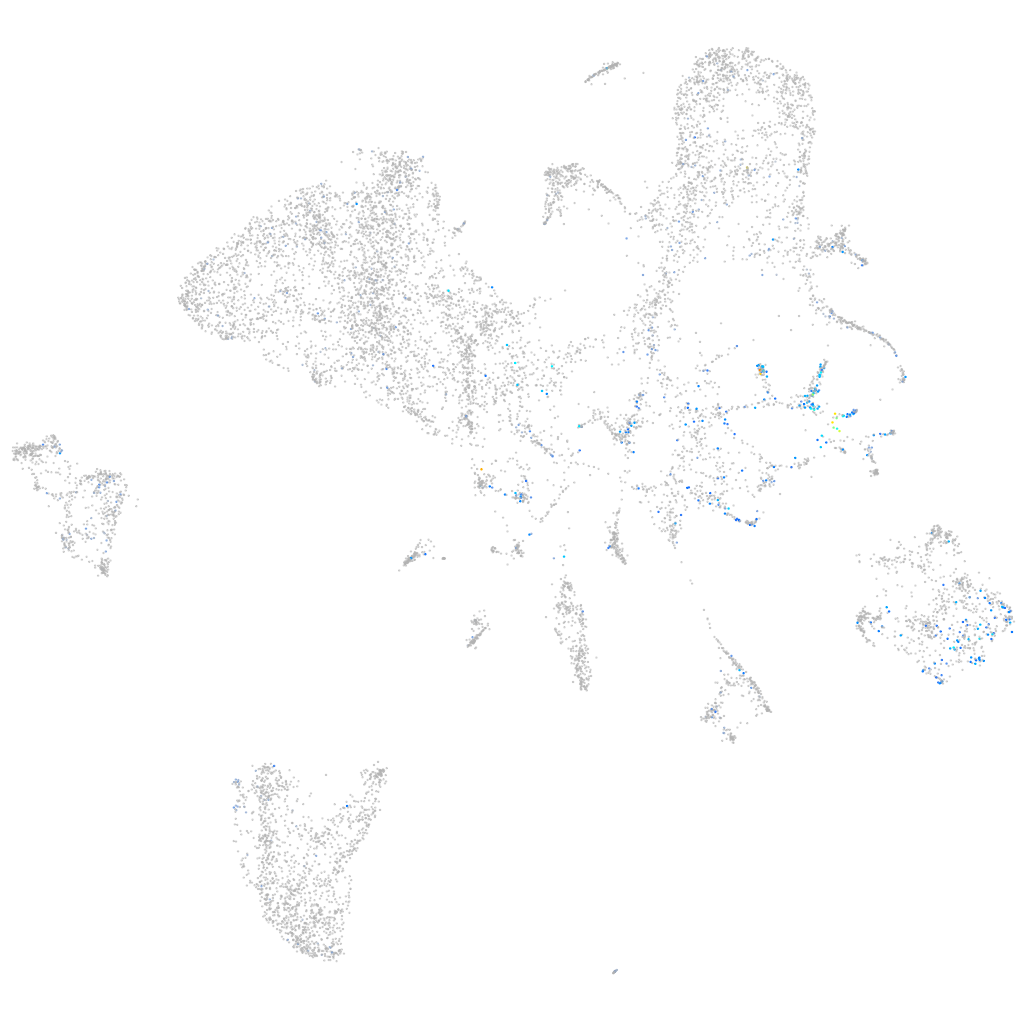
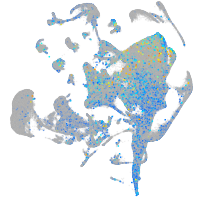
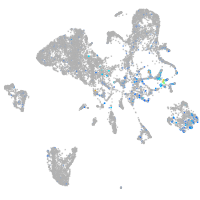
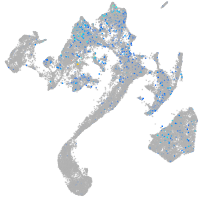
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| arxa | 0.258 | gamt | -0.149 |
| rprmb | 0.250 | ahcy | -0.148 |
| pax6b | 0.236 | gapdh | -0.145 |
| neurod1 | 0.229 | gatm | -0.134 |
| scg3 | 0.224 | bhmt | -0.121 |
| scgn | 0.218 | fbp1b | -0.121 |
| trmt9b | 0.215 | eno3 | -0.120 |
| kcnj11 | 0.211 | glud1b | -0.119 |
| c2cd4a | 0.206 | cx32.3 | -0.118 |
| ccka | 0.206 | mat1a | -0.117 |
| tspan7b | 0.198 | apoa4b.1 | -0.117 |
| pcsk1nl | 0.198 | apoa1b | -0.115 |
| bckdk | 0.198 | nupr1b | -0.115 |
| isl1 | 0.195 | apoc2 | -0.112 |
| insm1a | 0.187 | aldh7a1 | -0.111 |
| rasd4 | 0.187 | afp4 | -0.110 |
| zgc:92818 | 0.187 | scp2a | -0.109 |
| gfra3 | 0.186 | aldob | -0.108 |
| tspan7 | 0.186 | aldh6a1 | -0.105 |
| rem2 | 0.184 | gstt1a | -0.104 |
| ghrl | 0.182 | suclg1 | -0.103 |
| s100a1 | 0.181 | cdo1 | -0.103 |
| mir7a-1 | 0.180 | agxtb | -0.102 |
| si:dkey-153k10.9 | 0.180 | ugt1a7 | -0.102 |
| egr4 | 0.178 | dap | -0.101 |
| vat1 | 0.174 | mgst1.2 | -0.101 |
| marcksl1b | 0.173 | apoa2 | -0.101 |
| slc35g2b | 0.173 | apobb.1 | -0.101 |
| cacna1c | 0.171 | aqp12 | -0.100 |
| insm1b | 0.170 | suclg2 | -0.100 |
| kcnn3 | 0.169 | rdh1 | -0.100 |
| fev | 0.169 | acadm | -0.099 |
| chgb | 0.169 | gstp1 | -0.099 |
| lysmd2 | 0.169 | agxta | -0.098 |
| mir375-2 | 0.167 | grhprb | -0.097 |