DNA topoisomerase I mitochondrial
ZFIN 
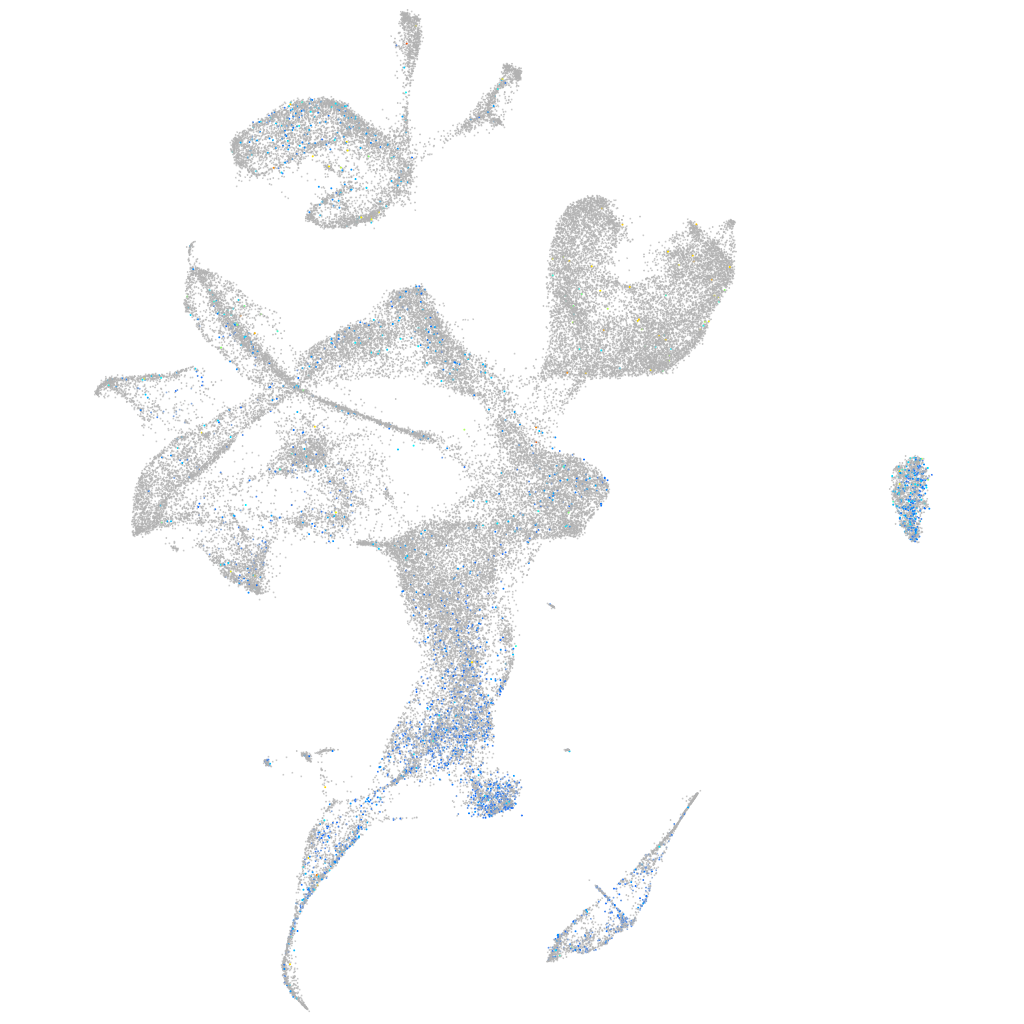
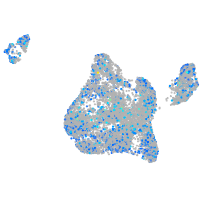
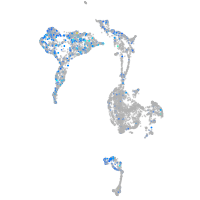
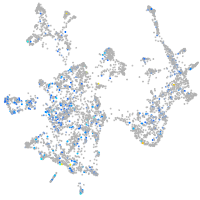
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| npm1a | 0.190 | ckbb | -0.109 |
| fbl | 0.170 | ptmaa | -0.100 |
| nop58 | 0.168 | CR383676.1 | -0.095 |
| dkc1 | 0.164 | gpm6aa | -0.087 |
| nop2 | 0.161 | syt5b | -0.080 |
| nhp2 | 0.160 | gpm6ab | -0.077 |
| pprc1 | 0.158 | pvalb1 | -0.076 |
| gnl3 | 0.155 | ptmab | -0.075 |
| snu13b | 0.154 | snap25b | -0.074 |
| nop56 | 0.153 | tuba1c | -0.074 |
| hspb1 | 0.153 | actc1b | -0.073 |
| ncl | 0.151 | rnasekb | -0.073 |
| rrp1 | 0.150 | pvalb2 | -0.072 |
| rsl1d1 | 0.150 | epb41a | -0.072 |
| mybbp1a | 0.149 | h3f3c | -0.071 |
| nop10 | 0.148 | cadm3 | -0.071 |
| si:ch211-217k17.7 | 0.147 | neurod4 | -0.070 |
| ebna1bp2 | 0.144 | sypb | -0.069 |
| aldob | 0.144 | atp6v0cb | -0.068 |
| bms1 | 0.144 | vsx1 | -0.065 |
| eif4ebp3l | 0.142 | crx | -0.064 |
| pes | 0.140 | gapdhs | -0.063 |
| prps1a | 0.140 | btg1 | -0.063 |
| wdr43 | 0.139 | calb2b | -0.061 |
| gar1 | 0.138 | rs1a | -0.061 |
| lyar | 0.138 | atp1b2a | -0.060 |
| ddx18 | 0.137 | foxg1b | -0.060 |
| si:dkey-102m7.3 | 0.136 | atpv0e2 | -0.060 |
| nip7 | 0.135 | fabp7a | -0.059 |
| tsr1 | 0.134 | mpp6b | -0.059 |
| rpf2 | 0.134 | hmgb1a | -0.059 |
| ddx27 | 0.134 | histh1l | -0.059 |
| llph | 0.134 | mylpfa | -0.058 |
| ddx24 | 0.134 | COX3 | -0.057 |
| paics | 0.132 | marcksl1a | -0.057 |