"troponin T type 3b (skeletal, fast)"
ZFIN 
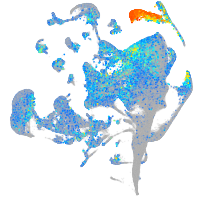
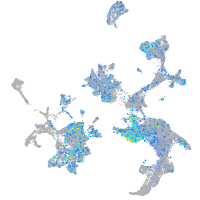
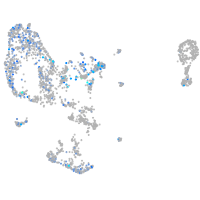
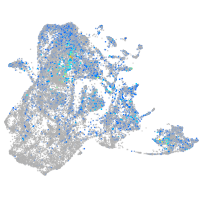
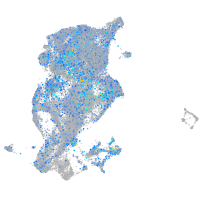
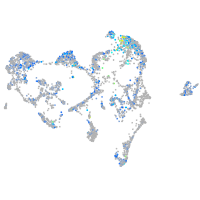
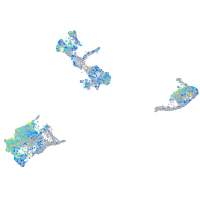
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pvalb2 | 0.379 | serbp1a | -0.150 |
| actc1b | 0.359 | ilf2 | -0.149 |
| pvalb1 | 0.358 | eif5a2 | -0.149 |
| mylpfa | 0.327 | si:ch211-222l21.1 | -0.148 |
| mylz3 | 0.302 | apex1 | -0.142 |
| mylpfb | 0.286 | seta | -0.141 |
| tnni2a.4 | 0.278 | snrpb | -0.139 |
| tpma | 0.273 | eif2s2 | -0.134 |
| apoa2 | 0.257 | zgc:194210 | -0.134 |
| ckma | 0.223 | nucks1a | -0.134 |
| myhz1.1 | 0.211 | zbtb16a | -0.133 |
| acta1b | 0.207 | ddx39ab | -0.133 |
| nme2b.2 | 0.205 | rps12 | -0.132 |
| ak1 | 0.203 | oc90 | -0.130 |
| CELA1 (1 of many) | 0.201 | ptges3b | -0.129 |
| apoa1b | 0.201 | sub1a | -0.127 |
| prss1 | 0.185 | CABZ01075068.1 | -0.126 |
| prss59.2 | 0.184 | snrpd1 | -0.125 |
| pvalb4 | 0.183 | snrpe | -0.125 |
| ttn.2 | 0.178 | snrpg | -0.122 |
| myhz2 | 0.175 | hnrnph1l | -0.121 |
| gapdh | 0.172 | hmga1a | -0.119 |
| hbbe1.1 | 0.167 | igf2bp1 | -0.119 |
| ttn.1 | 0.165 | snrpf | -0.119 |
| ctrb1 | 0.164 | snu13b | -0.118 |
| krt17 | 0.163 | syncrip | -0.117 |
| ckmb | 0.161 | khdrbs1a | -0.117 |
| ela3l | 0.153 | anp32b | -0.117 |
| icn2 | 0.151 | ppid | -0.117 |
| myl1 | 0.151 | marcksb | -0.117 |
| cyt1 | 0.150 | ppp2cb | -0.116 |
| tcnba | 0.145 | sae1 | -0.115 |
| CR383676.1 | 0.144 | snrpd2 | -0.115 |
| zgc:112160 | 0.143 | atp5if1a | -0.115 |
| myhz1.2 | 0.143 | chaf1a | -0.115 |