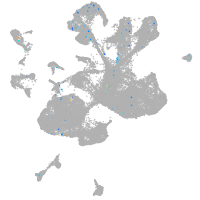
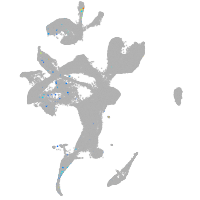
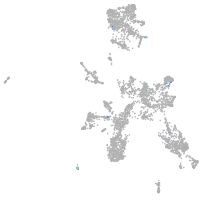
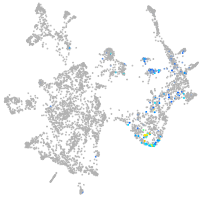
"tumor necrosis factor receptor superfamily, member 9a"
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| slc45a2 | 0.127 | hmgb2a | -0.057 |
| tyrp1a | 0.126 | hmga1a | -0.045 |
| slc24a5 | 0.124 | ptmab | -0.042 |
| agtrap | 0.124 | stmn1a | -0.041 |
| pmelb | 0.123 | hmgb2b | -0.040 |
| oca2 | 0.122 | fabp7a | -0.037 |
| fabp11b | 0.119 | rrm1 | -0.036 |
| tyr | 0.119 | mki67 | -0.036 |
| gpr143 | 0.118 | pcna | -0.036 |
| dct | 0.116 | si:ch73-1a9.3 | -0.034 |
| pnp4a | 0.116 | ccna2 | -0.034 |
| tyrp1b | 0.115 | si:ch211-222l21.1 | -0.033 |
| hps1 | 0.110 | dut | -0.033 |
| pah | 0.108 | banf1 | -0.033 |
| zgc:110591 | 0.108 | nasp | -0.032 |
| pmela | 0.107 | si:ch211-137a8.4 | -0.032 |
| rab32a | 0.106 | rrm2 | -0.031 |
| mb | 0.106 | dek | -0.031 |
| si:ch73-86n18.1 | 0.105 | hes6 | -0.031 |
| rab38 | 0.103 | ncapg | -0.031 |
| msnb | 0.103 | fbxo5 | -0.031 |
| cracr2ab | 0.103 | ppm1g | -0.030 |
| mitfa | 0.102 | si:dkey-261m9.17 | -0.030 |
| tspan10 | 0.101 | zgc:110425 | -0.030 |
| tspan36 | 0.101 | lig1 | -0.029 |
| tm6sf2 | 0.100 | hmgn2 | -0.029 |
| nog3 | 0.098 | chaf1a | -0.029 |
| fgfr3 | 0.096 | zgc:110540 | -0.029 |
| bace2 | 0.096 | ran | -0.029 |
| qdpra | 0.094 | ccnd1 | -0.029 |
| FP085398.1 | 0.093 | cx43.4 | -0.029 |
| fgfbp1a | 0.093 | lbr | -0.029 |
| rgrb | 0.091 | h2afva | -0.029 |
| stra6 | 0.091 | nutf2l | -0.029 |
| gstp1 | 0.090 | rpa3 | -0.029 |