thymosin beta 2
ZFIN 
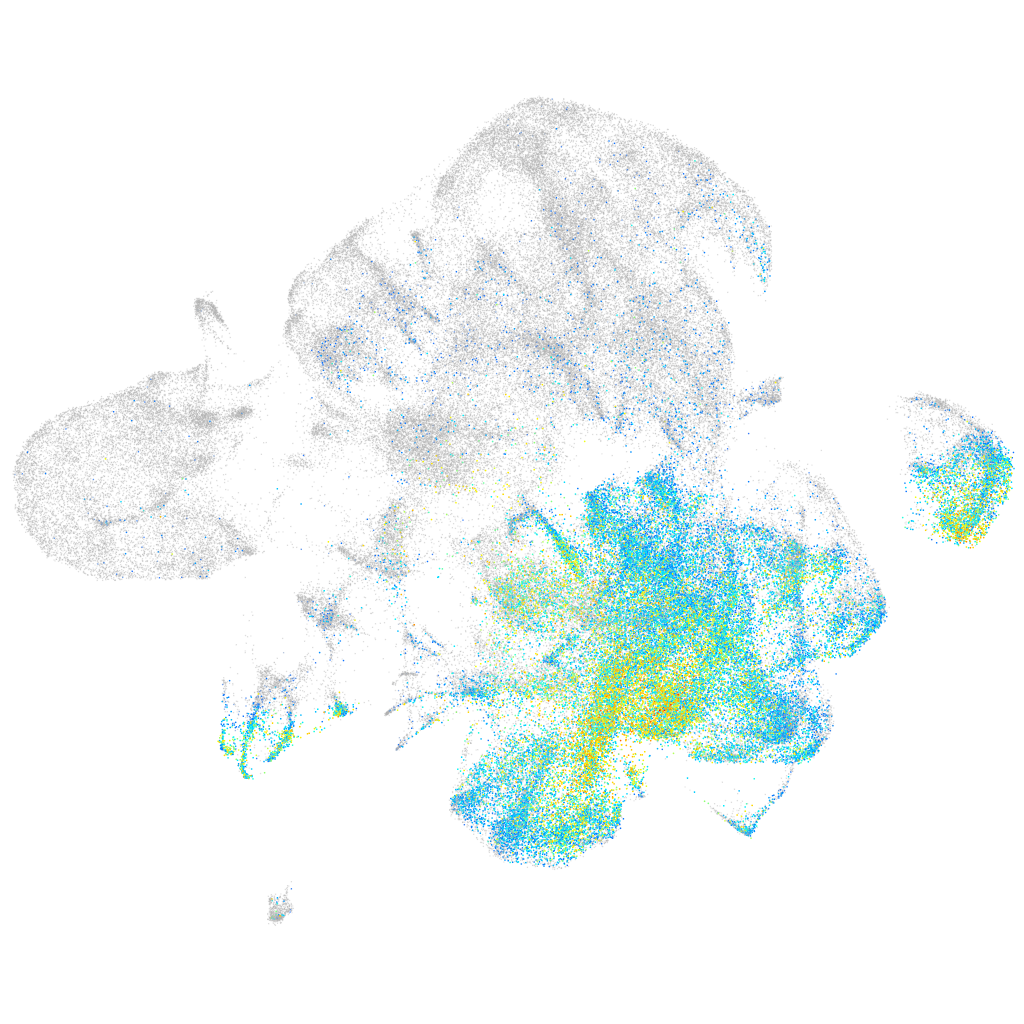
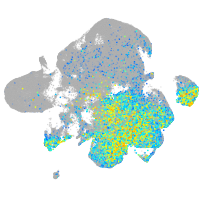
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| stmn2a | 0.612 | hmgb2a | -0.513 |
| sncb | 0.603 | hmga1a | -0.454 |
| gng3 | 0.596 | hmgb2b | -0.423 |
| gap43 | 0.596 | id1 | -0.386 |
| ywhag2 | 0.589 | pcna | -0.377 |
| zgc:65894 | 0.573 | stmn1a | -0.365 |
| rtn1b | 0.557 | mdka | -0.354 |
| elavl4 | 0.545 | XLOC-003690 | -0.348 |
| snap25a | 0.545 | eef1a1l1 | -0.347 |
| mllt11 | 0.533 | si:dkey-151g10.6 | -0.347 |
| atp6v0cb | 0.533 | her15.1 | -0.347 |
| stmn1b | 0.521 | msna | -0.340 |
| gapdhs | 0.520 | rplp2l | -0.336 |
| stxbp1a | 0.517 | rps12 | -0.335 |
| tuba1c | 0.513 | rplp1 | -0.335 |
| stx1b | 0.512 | si:ch73-281n10.2 | -0.330 |
| atp6v1e1b | 0.508 | XLOC-003689 | -0.329 |
| gpm6ab | 0.506 | anp32b | -0.328 |
| ckbb | 0.506 | sox3 | -0.326 |
| vamp2 | 0.505 | ranbp1 | -0.324 |
| atp6v1g1 | 0.497 | chaf1a | -0.321 |
| calm1a | 0.493 | selenoh | -0.320 |
| calm2a | 0.487 | ran | -0.318 |
| maptb | 0.480 | sox19a | -0.316 |
| rtn1a | 0.466 | rps20 | -0.316 |
| tpi1b | 0.464 | rrm1 | -0.316 |
| rnasekb | 0.461 | ccnd1 | -0.314 |
| map1aa | 0.458 | tuba8l | -0.314 |
| zgc:153426 | 0.457 | tspan7 | -0.313 |
| rab6bb | 0.454 | mcm7 | -0.313 |
| id4 | 0.450 | mki67 | -0.309 |
| tuba2 | 0.443 | npm1a | -0.309 |
| sncgb | 0.440 | rpsa | -0.308 |
| ywhag1 | 0.439 | XLOC-003692 | -0.305 |
| si:dkeyp-75h12.5 | 0.439 | banf1 | -0.305 |