transmembrane protein 9
ZFIN 
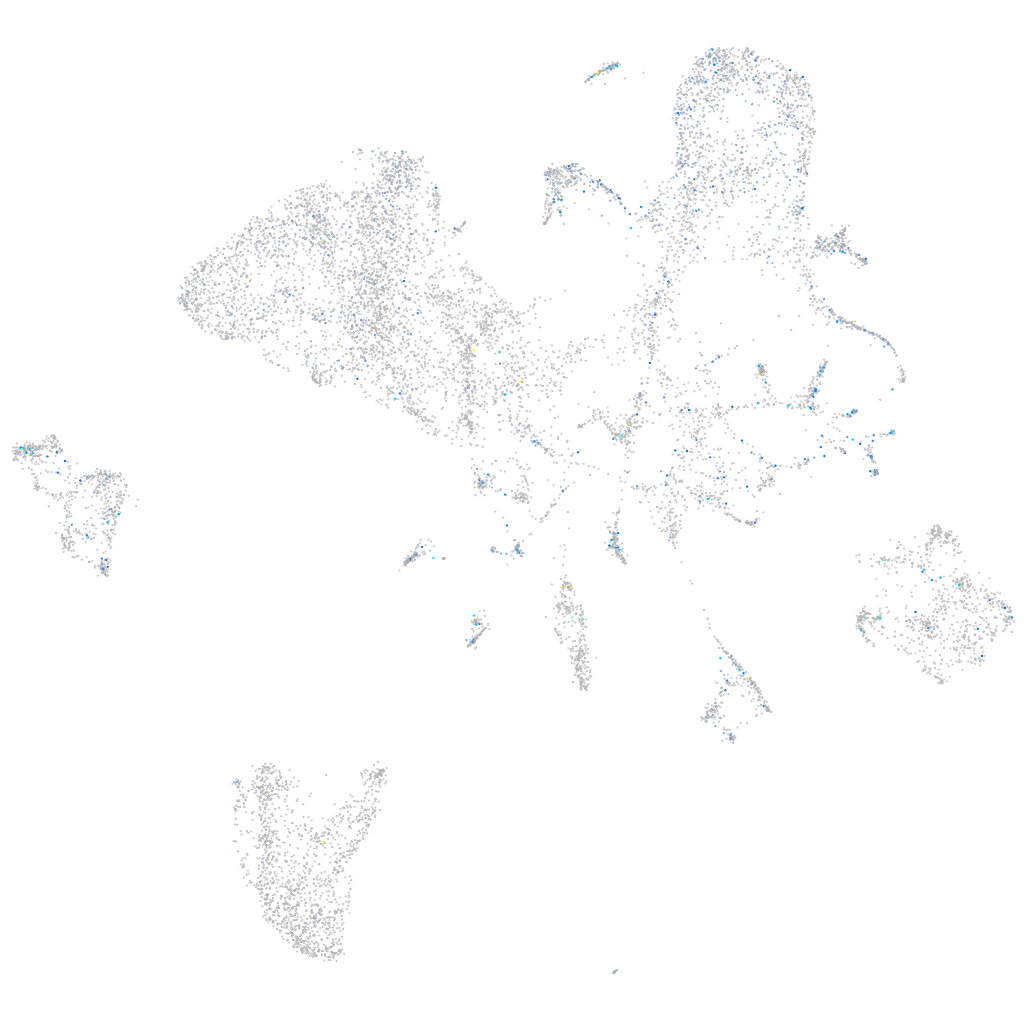
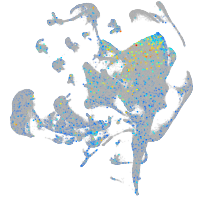
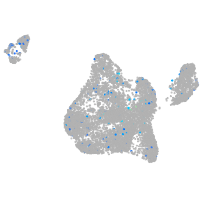
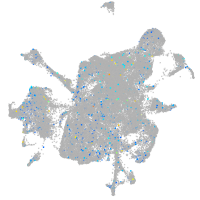
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.137 | bhmt | -0.099 |
| si:ch211-137i24.10 | 0.130 | aqp12 | -0.093 |
| arpc3 | 0.125 | agxtb | -0.079 |
| sat1a.2 | 0.124 | mat1a | -0.077 |
| degs2 | 0.121 | gatm | -0.077 |
| ptchd1 | 0.121 | ppdpfa | -0.077 |
| cldn15la | 0.115 | fabp3 | -0.075 |
| XLOC-026336 | 0.114 | gamt | -0.074 |
| cd63 | 0.114 | apoc1 | -0.073 |
| vil1 | 0.114 | prss1 | -0.071 |
| ier2b | 0.112 | fabp10a | -0.069 |
| rims2a | 0.112 | cel.2 | -0.069 |
| sri | 0.112 | ela2l | -0.069 |
| tm4sf4 | 0.112 | prss59.2 | -0.069 |
| elovl1b | 0.112 | cel.1 | -0.068 |
| slc3a2a | 0.111 | prss59.1 | -0.068 |
| adra1ba | 0.111 | zgc:112160 | -0.068 |
| cap1 | 0.111 | CELA1 (1 of many) | -0.068 |
| nipa2 | 0.111 | cpa5 | -0.068 |
| calm2a | 0.111 | cpb1 | -0.067 |
| neurod1 | 0.110 | hao1 | -0.067 |
| trpm3 | 0.110 | ela3l | -0.067 |
| clta | 0.110 | ttc36 | -0.066 |
| si:dkeyp-72e1.6 | 0.110 | rbp2b | -0.066 |
| cst14a.2 | 0.110 | cpa1 | -0.066 |
| GCA | 0.110 | tfa | -0.066 |
| lrrc3b | 0.110 | sycn.2 | -0.065 |
| atp6ap2 | 0.109 | ctrb1 | -0.065 |
| gfra3 | 0.109 | ctrl | -0.065 |
| lysmd2 | 0.109 | fep15 | -0.065 |
| calm1b | 0.108 | apoa2 | -0.065 |
| ppiab | 0.108 | apoda.2 | -0.065 |
| tmem50a | 0.107 | si:ch211-240l19.5 | -0.065 |
| tmem176l.2 | 0.107 | zgc:136461 | -0.065 |
| gnb1a | 0.107 | cpa4 | -0.064 |