transmembrane protein 56b [Source:ZFIN;Acc:ZDB-GENE-041010-90]
ZFIN 
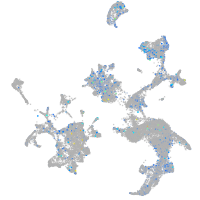
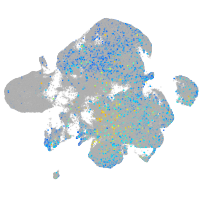
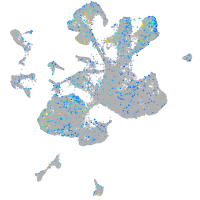
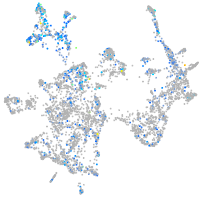
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gapdhs | 0.125 | si:dkey-151g10.6 | -0.114 |
| snap25a | 0.124 | rplp1 | -0.110 |
| ywhag2 | 0.123 | rpl23 | -0.105 |
| atp6v0cb | 0.123 | rps20 | -0.099 |
| stmn2a | 0.122 | rps19 | -0.097 |
| cplx2l | 0.120 | rps15a | -0.095 |
| sypb | 0.119 | rpl37 | -0.094 |
| rbfox1 | 0.115 | hmgb2a | -0.093 |
| stx1b | 0.115 | rps24 | -0.092 |
| stxbp1a | 0.114 | rpl21 | -0.092 |
| ckbb | 0.113 | rpl39 | -0.092 |
| kcnc3a | 0.113 | rps7 | -0.091 |
| tmem59l | 0.113 | rpl11 | -0.091 |
| sncgb | 0.113 | rpl10a | -0.091 |
| elavl4 | 0.111 | rps14 | -0.090 |
| nptna | 0.111 | XLOC-003689 | -0.089 |
| cplx2 | 0.109 | rpl27 | -0.089 |
| rnasekb | 0.109 | hmgb2b | -0.089 |
| aldocb | 0.108 | rps27a | -0.089 |
| gjd1a | 0.107 | rpl7 | -0.089 |
| gng3 | 0.106 | rpl12 | -0.089 |
| eno2 | 0.106 | cx43.4 | -0.088 |
| cpe | 0.105 | rplp2l | -0.088 |
| rtn1b | 0.105 | rps27.1 | -0.087 |
| sypa | 0.105 | rpl26 | -0.087 |
| si:ch73-119p20.1 | 0.105 | rps2 | -0.087 |
| grin1a | 0.104 | rps12 | -0.087 |
| syt2a | 0.104 | rpl13 | -0.086 |
| sncb | 0.103 | rps25 | -0.084 |
| atp2b3a | 0.103 | snrpf | -0.083 |
| map1aa | 0.103 | mcm7 | -0.083 |
| calm1b | 0.103 | rps13 | -0.083 |
| zgc:65894 | 0.103 | rps28 | -0.082 |
| nsg2 | 0.103 | rpl17 | -0.082 |
| calm3a | 0.102 | rpl35a | -0.081 |