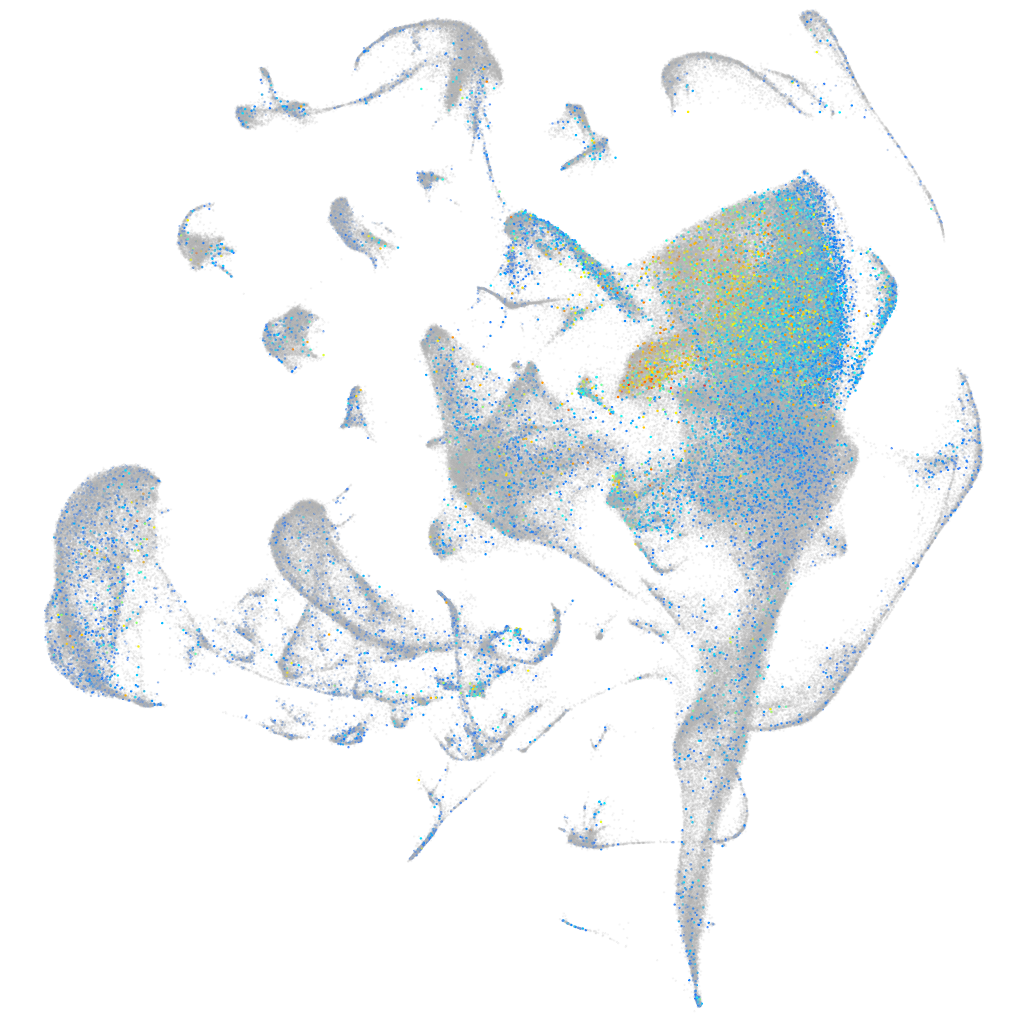transmembrane protein 53
ZFIN 
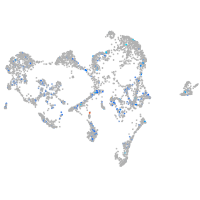
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| elavl3 | 0.156 | aldob | -0.075 |
| stmn1b | 0.155 | eif4ebp3l | -0.069 |
| tuba1c | 0.150 | ccng1 | -0.068 |
| rtn1a | 0.148 | id1 | -0.068 |
| tubb5 | 0.147 | npm1a | -0.068 |
| gpm6ab | 0.141 | hspb1 | -0.066 |
| tmeff1b | 0.138 | sdc4 | -0.063 |
| fez1 | 0.136 | bzw1b | -0.062 |
| gpm6aa | 0.136 | fosab | -0.061 |
| nova2 | 0.136 | hdlbpa | -0.061 |
| zc4h2 | 0.136 | vamp3 | -0.060 |
| si:dkey-276j7.1 | 0.135 | dkc1 | -0.059 |
| ywhah | 0.132 | nop58 | -0.059 |
| hmgb3a | 0.131 | rsl1d1 | -0.059 |
| myt1b | 0.131 | eef1da | -0.058 |
| gng3 | 0.129 | akap12b | -0.057 |
| tmsb | 0.129 | atp1b1a | -0.057 |
| tuba1a | 0.128 | banf1 | -0.057 |
| chd4a | 0.126 | fbl | -0.057 |
| sox4a | 0.126 | selenoh | -0.057 |
| celf3a | 0.125 | pcna | -0.056 |
| ckbb | 0.125 | sparc | -0.054 |
| gng2 | 0.125 | ccnd1 | -0.053 |
| myt1a | 0.123 | chaf1a | -0.053 |
| dpysl3 | 0.122 | lig1 | -0.053 |
| dpysl2b | 0.122 | nop10 | -0.053 |
| nsg2 | 0.120 | slc38a5b | -0.053 |
| hnrnpa0a | 0.119 | apoc1 | -0.052 |
| celf2 | 0.117 | snu13b | -0.052 |
| jagn1a | 0.117 | ahnak | -0.051 |
| scrt2 | 0.117 | cdca7a | -0.051 |
| rtn1b | 0.116 | nop56 | -0.051 |
| adam22 | 0.115 | si:ch211-152c2.3 | -0.051 |
| epb41a | 0.115 | si:dkey-16p21.8 | -0.051 |
| csdc2a | 0.114 | stm | -0.051 |