transmembrane protein 119b
ZFIN 
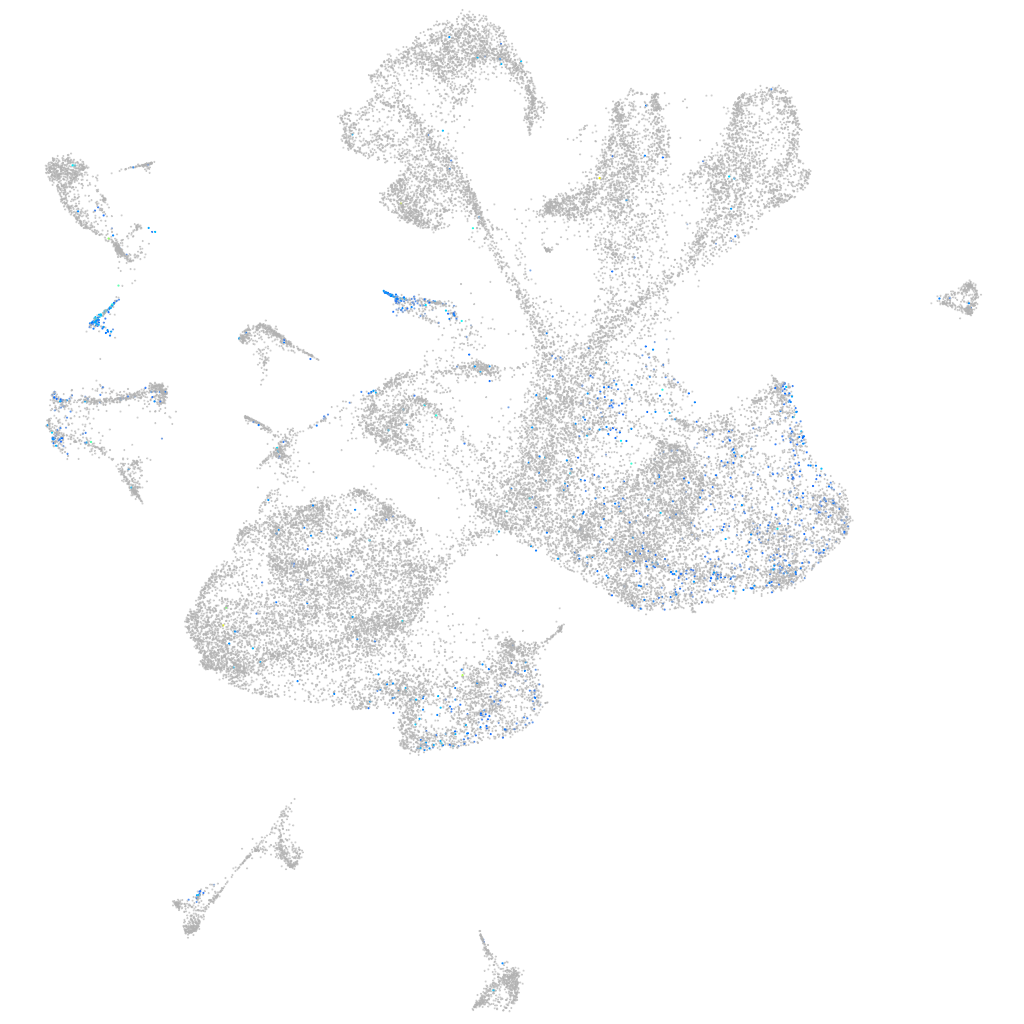
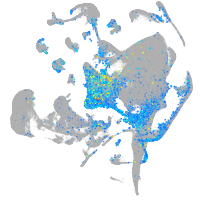
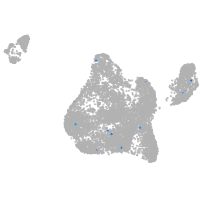
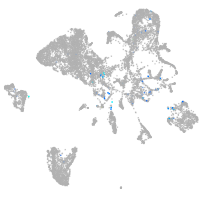
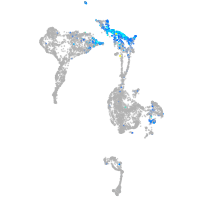
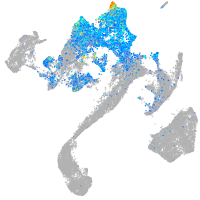
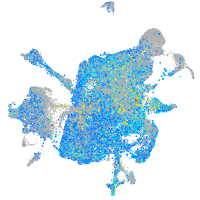
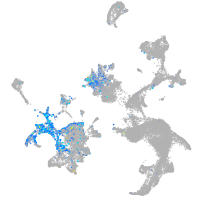
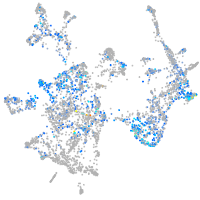
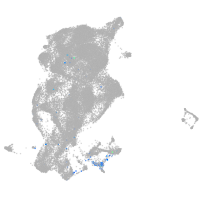
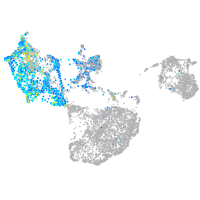
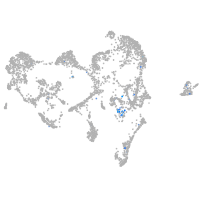
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch211-157b11.8 | 0.219 | ckbb | -0.106 |
| twist2 | 0.190 | gpm6aa | -0.103 |
| ntd5 | 0.180 | tuba1c | -0.085 |
| apela | 0.158 | rtn1a | -0.084 |
| lamb1a | 0.157 | nova2 | -0.083 |
| spon2a | 0.156 | CU467822.1 | -0.080 |
| ednrab | 0.155 | pvalb2 | -0.079 |
| fkbp7 | 0.155 | pvalb1 | -0.075 |
| timp4.1 | 0.154 | myt1b | -0.074 |
| XLOC-042222 | 0.153 | fabp7a | -0.074 |
| ptx3a | 0.153 | gpm6ab | -0.074 |
| AL954327.1 | 0.150 | tmsb | -0.070 |
| col8a1a | 0.146 | gnao1a | -0.069 |
| inka1a | 0.145 | ptmaa | -0.069 |
| kng1 | 0.145 | stmn1b | -0.068 |
| crestin | 0.144 | COX3 | -0.068 |
| fbn2b | 0.137 | hbbe1.3 | -0.068 |
| gdf6b | 0.135 | elavl3 | -0.067 |
| plod2 | 0.134 | elavl4 | -0.067 |
| serpinh1b | 0.134 | slc1a2b | -0.066 |
| pgfb | 0.133 | rnasekb | -0.066 |
| optc | 0.133 | dpysl2b | -0.066 |
| aplnr2 | 0.131 | CU634008.1 | -0.065 |
| XLOC-001964 | 0.131 | hmgb1a | -0.064 |
| cdx4 | 0.131 | actc1b | -0.064 |
| matn3b | 0.130 | gng3 | -0.064 |
| angpt1 | 0.129 | fez1 | -0.063 |
| mfap2 | 0.127 | hbae3 | -0.063 |
| krt18a.1 | 0.127 | atp1a1b | -0.063 |
| fgfrl1b | 0.126 | gapdhs | -0.062 |
| colec12 | 0.126 | vamp2 | -0.061 |
| tfec | 0.125 | CR848047.1 | -0.061 |
| eif4ebp3l | 0.120 | si:dkeyp-75h12.5 | -0.060 |
| tpm4a | 0.120 | tmsb4x | -0.059 |
| gdf6a | 0.117 | cadm3 | -0.058 |