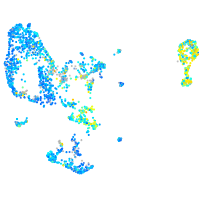translation machinery associated 7 homolog
ZFIN 
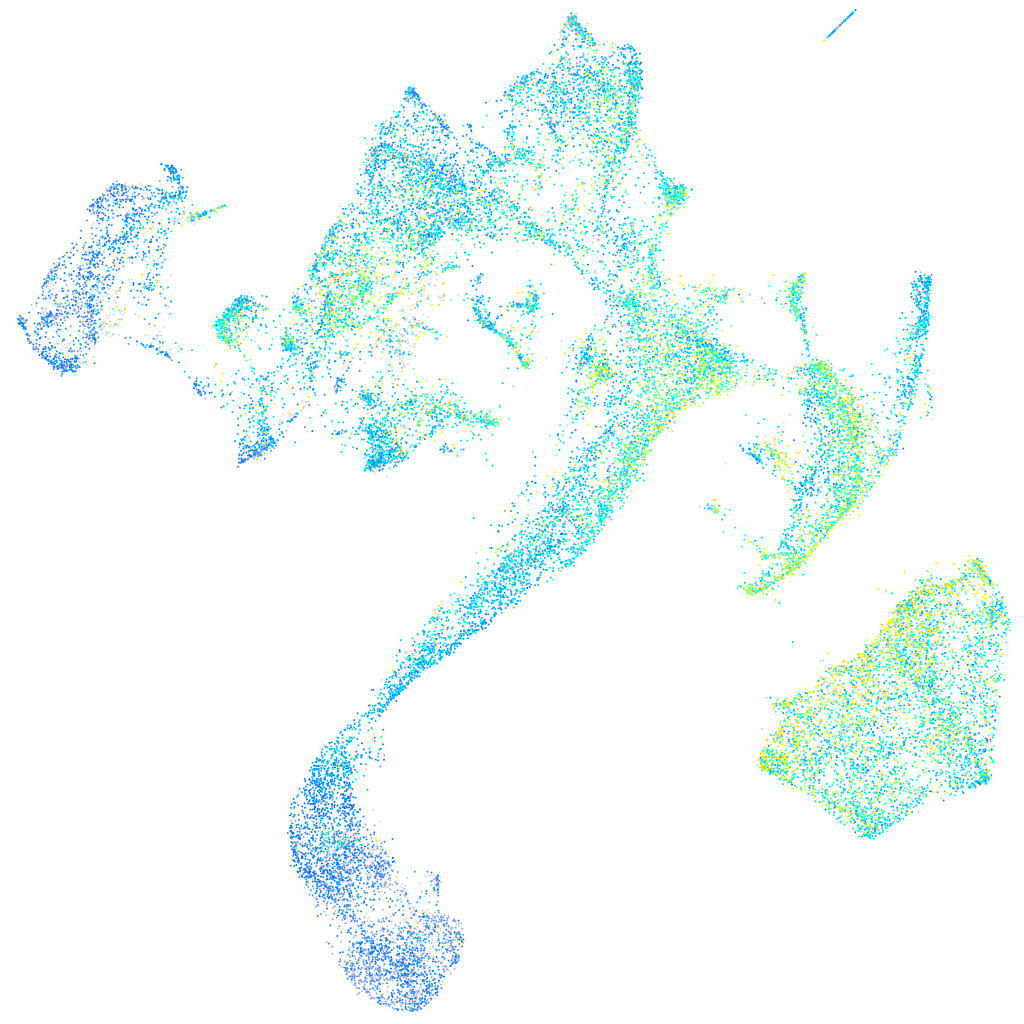
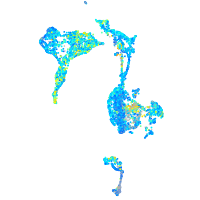
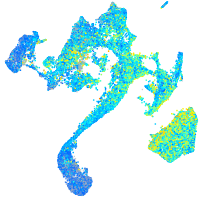
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| h2afvb | 0.671 | ckma | -0.556 |
| hmga1a | 0.652 | actc1b | -0.553 |
| hmgb2b | 0.640 | ak1 | -0.548 |
| snrpf | 0.636 | ckmb | -0.546 |
| cirbpa | 0.635 | neb | -0.541 |
| hnrnpabb | 0.634 | atp2a1 | -0.535 |
| hnrnpaba | 0.629 | actn3a | -0.534 |
| khdrbs1a | 0.625 | tnnc2 | -0.530 |
| setb | 0.617 | si:ch73-367p23.2 | -0.530 |
| hmgb2a | 0.609 | nme2b.2 | -0.522 |
| snrpe | 0.608 | gapdh | -0.521 |
| snrpd1 | 0.606 | smyd1a | -0.516 |
| snrpb | 0.605 | ldb3b | -0.515 |
| cbx3a | 0.601 | pvalb1 | -0.514 |
| snrpd2 | 0.597 | myom1a | -0.513 |
| ran | 0.596 | actn3b | -0.508 |
| snrpg | 0.595 | aldoab | -0.508 |
| h3f3d | 0.591 | si:ch211-266g18.10 | -0.508 |
| snu13b | 0.585 | pkmb | -0.507 |
| si:ch73-281n10.2 | 0.583 | eno3 | -0.504 |
| hmgn2 | 0.582 | XLOC-025819 | -0.495 |
| npm1a | 0.581 | si:dkey-16p21.8 | -0.494 |
| ddx39ab | 0.581 | pvalb2 | -0.494 |
| hmgn7 | 0.578 | ttn.2 | -0.492 |
| rbm8a | 0.576 | pgam2 | -0.490 |
| hnrnpa0b | 0.573 | si:ch211-255p10.3 | -0.490 |
| pabpc1a | 0.572 | prx | -0.490 |
| ptges3b | 0.565 | tmod4 | -0.489 |
| si:ch211-222l21.1 | 0.563 | mylpfa | -0.485 |
| nop58 | 0.557 | casq1b | -0.482 |
| cirbpb | 0.556 | XLOC-001975 | -0.479 |
| ncl | 0.555 | acta1b | -0.479 |
| syncrip | 0.555 | tnnt3b | -0.479 |
| anp32b | 0.554 | mylz3 | -0.479 |
| si:ch73-1a9.3 | 0.553 | ank1a | -0.478 |