tyrosine kinase with immunoglobulin-like and EGF-like domains 1
ZFIN 
Other cell groups


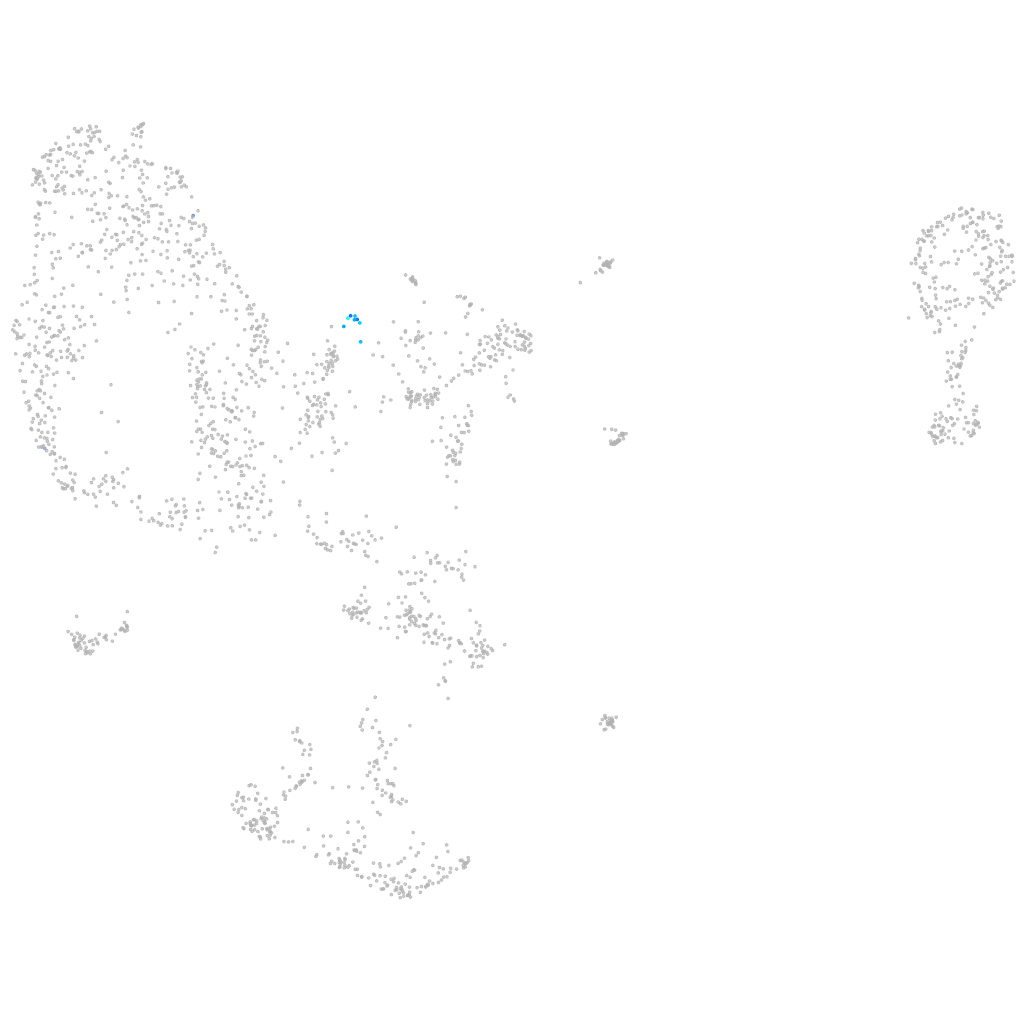
all cells |
blastomeres |
PGCs |
endoderm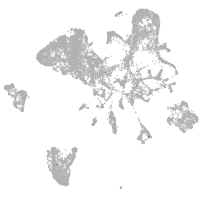 |
axial mesoderm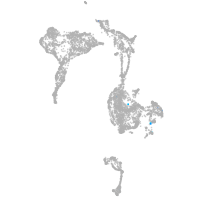 |
muscle  |
mural cells / non-skeletal muscle 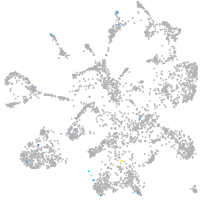 |
mesenchyme 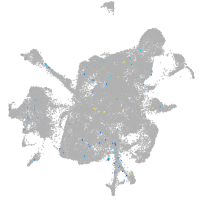 |
hematopoietic / vasculature 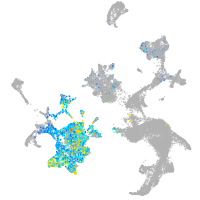 |
pronephros 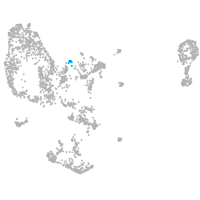 |
neural |
spinal cord / glia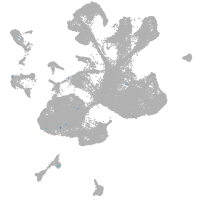 |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch211-145b13.6 | 0.826 | rps8a | -0.075 |
| cdh5 | 0.821 | eif2s2 | -0.065 |
| plvapb | 0.744 | rpl17 | -0.064 |
| pecam1 | 0.707 | hnrnpaba | -0.063 |
| stab1 | 0.698 | si:ch73-281n10.2 | -0.053 |
| si:dkeyp-97a10.2 | 0.694 | hmgn7 | -0.053 |
| gpr182 | 0.679 | fth1a | -0.049 |
| kdrl | 0.672 | sub1a | -0.048 |
| myct1a | 0.654 | sap18 | -0.048 |
| XLOC-043043 | 0.619 | f11r.1 | -0.048 |
| kdr | 0.614 | tma7 | -0.048 |
| agtr1b | 0.611 | pax2a | -0.047 |
| si:ch211-14k19.8 | 0.601 | ilf2 | -0.047 |
| lyve1a | 0.599 | rack1 | -0.046 |
| ptprb | 0.590 | hnf1ba | -0.046 |
| aqp7 | 0.583 | hmga1a | -0.046 |
| ecscr | 0.579 | ptges3b | -0.046 |
| fgd5a | 0.568 | serbp1a | -0.045 |
| myct1b | 0.560 | tomm22 | -0.044 |
| calcrlb | 0.551 | cldnc | -0.043 |
| loxl1 | 0.545 | ccdc47 | -0.043 |
| ptger1b | 0.544 | psma5 | -0.043 |
| adra1d | 0.538 | cldnh | -0.043 |
| si:ch211-33e4.2 | 0.519 | eif3ba | -0.043 |
| hspa12b | 0.518 | top1l | -0.043 |
| mrc1a | 0.515 | hoxb3a | -0.042 |
| sox18 | 0.498 | zgc:193541 | -0.042 |
| arhgap31 | 0.487 | nop58 | -0.042 |
| rca2.2 | 0.487 | hnrnpd | -0.041 |
| capn5a | 0.481 | eif3g | -0.041 |
| flt1 | 0.480 | snrpf | -0.041 |
| si:ch73-334d15.2 | 0.480 | drap1 | -0.041 |
| plvapa | 0.479 | mrpl57 | -0.041 |
| arhgef9b | 0.472 | si:ch211-288g17.3 | -0.041 |
| clec14a | 0.470 | khdrbs1a | -0.040 |