TIAM Rac1 associated GEF 1a
ZFIN 
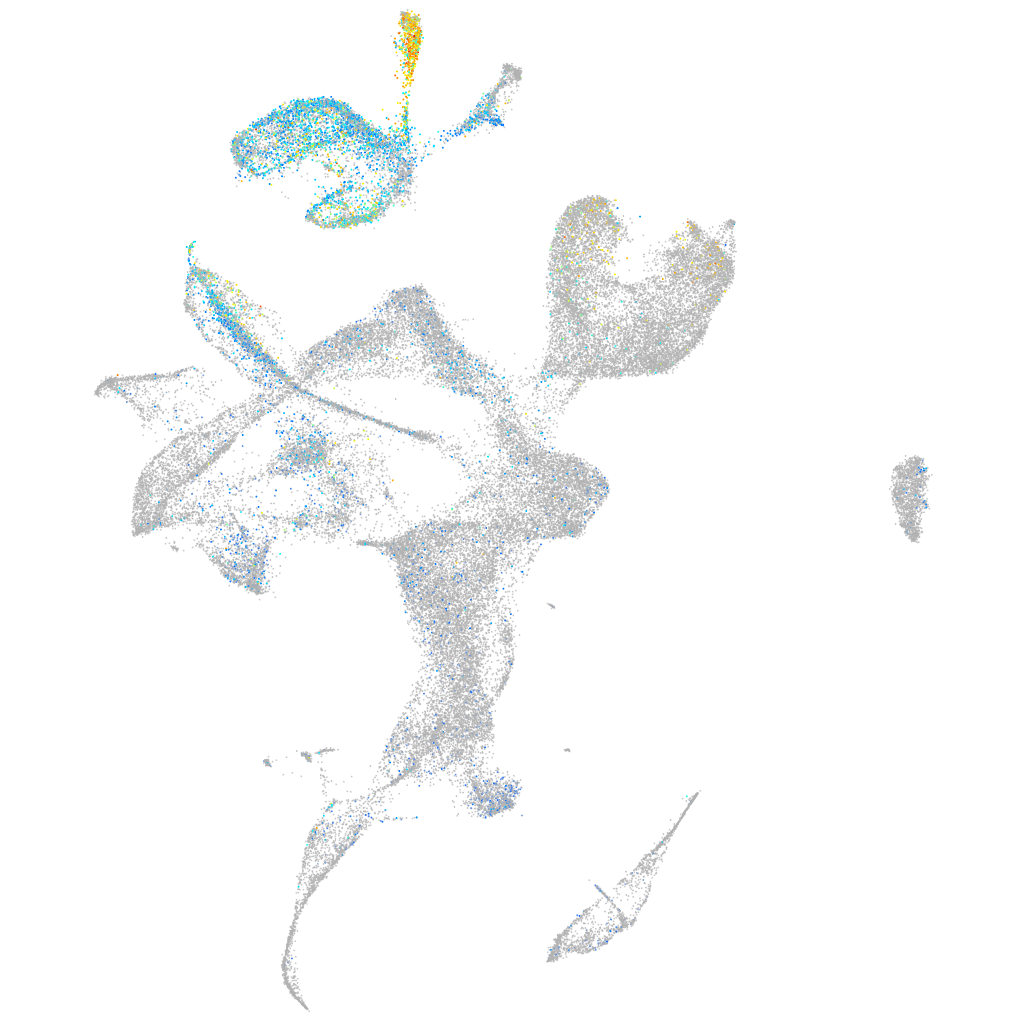
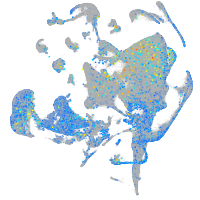
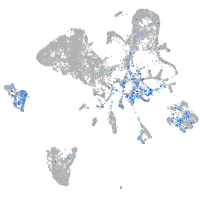
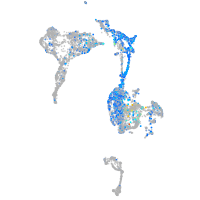
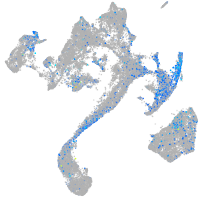
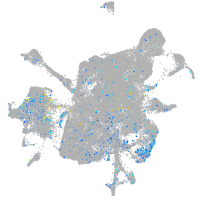
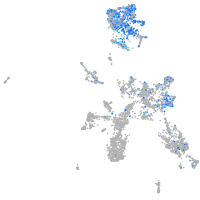
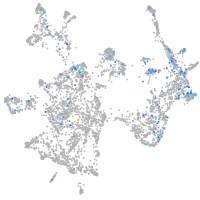
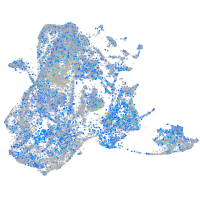
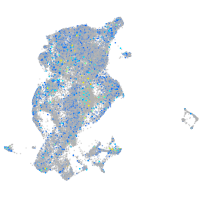
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| slc32a1 | 0.389 | hmgb2a | -0.249 |
| CR855337.1 | 0.384 | msna | -0.147 |
| tfap2a | 0.382 | pcna | -0.140 |
| pvalb6 | 0.374 | tuba8l4 | -0.138 |
| gldn | 0.357 | msi1 | -0.136 |
| syt1a | 0.348 | rrm1 | -0.128 |
| slc18a3a | 0.340 | stmn1a | -0.128 |
| rbfox1 | 0.337 | eef1da | -0.127 |
| stxbp1a | 0.335 | ahcy | -0.127 |
| elavl3 | 0.328 | dut | -0.126 |
| mir181b-3 | 0.315 | tuba8l | -0.125 |
| stx1b | 0.305 | her15.1 | -0.125 |
| tfap2b | 0.305 | nutf2l | -0.124 |
| pax6b | 0.297 | mki67 | -0.124 |
| LOC101885188 | 0.297 | hmga1a | -0.122 |
| rgs9bp | 0.296 | crx | -0.121 |
| pax10 | 0.292 | selenoh | -0.120 |
| barhl2 | 0.287 | banf1 | -0.120 |
| ywhag2 | 0.279 | ccna2 | -0.119 |
| nrxn1a | 0.278 | lbr | -0.118 |
| plppr5a | 0.277 | ccnd1 | -0.118 |
| si:ch73-119p20.1 | 0.276 | mcm7 | -0.116 |
| gabrb4 | 0.273 | chaf1a | -0.116 |
| lrrn2 | 0.272 | id1 | -0.116 |
| gpr158a | 0.269 | mdka | -0.115 |
| rbfox2 | 0.265 | rpa3 | -0.114 |
| pax6a | 0.264 | neurod4 | -0.113 |
| stmn1b | 0.263 | otx5 | -0.113 |
| st8sia5 | 0.263 | dek | -0.112 |
| gnb1a | 0.259 | tubb4b | -0.111 |
| slc6a1a | 0.258 | rrm2 | -0.110 |
| si:ch73-290k24.5 | 0.258 | lig1 | -0.110 |
| megf10 | 0.255 | cks1b | -0.110 |
| pcdh19 | 0.255 | ddah2 | -0.109 |
| gnao1a | 0.252 | syt5b | -0.108 |