thymocyte selection associated family member 2
ZFIN 
Other cell groups


all cells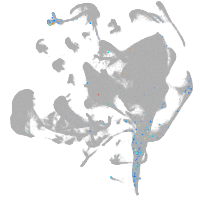 |
blastomeres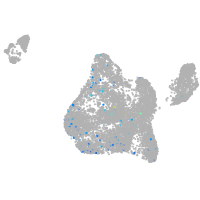 |
PGCs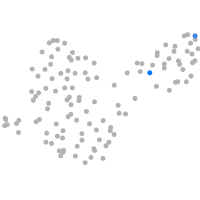 |
endoderm |
axial mesoderm |
muscle 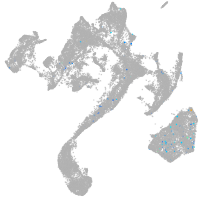 |
mural cells / non-skeletal muscle  |
mesenchyme 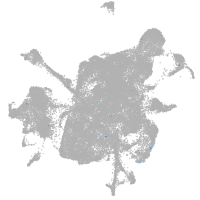 |
hematopoietic / vasculature 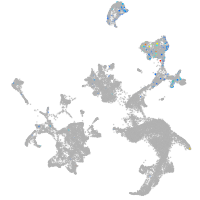 |
pronephros 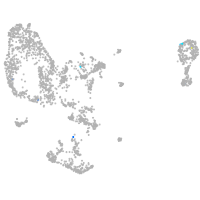 |
neural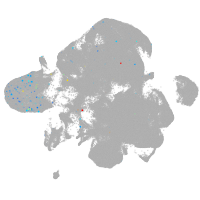 |
spinal cord / glia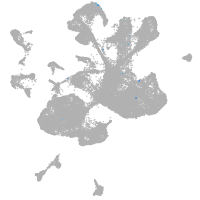 |
eye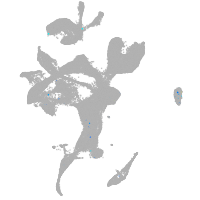 |
taste / olfactory |
otic / lateral line |
epidermis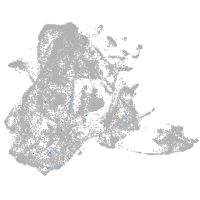 |
periderm |
fin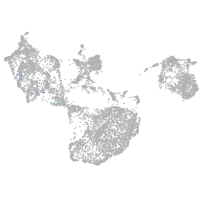 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| spi1b | 0.126 | gpm6aa | -0.029 |
| fcer1gl | 0.122 | krt4 | -0.028 |
| laptm5 | 0.122 | tuba1c | -0.028 |
| fcer1g | 0.119 | cyt1 | -0.027 |
| ptprc | 0.117 | fabp3 | -0.025 |
| zgc:64051 | 0.116 | hbbe1.3 | -0.025 |
| myo1f | 0.115 | pvalb1 | -0.025 |
| si:ch211-102c2.4 | 0.114 | rnasekb | -0.025 |
| coro1a | 0.113 | rtn1a | -0.025 |
| LOC100536213 | 0.113 | cyt1l | -0.024 |
| FERMT3 (1 of many) | 0.113 | gpm6ab | -0.024 |
| itgb2 | 0.112 | pvalb2 | -0.024 |
| lcp1 | 0.112 | calm1a | -0.024 |
| CABZ01073834.1 | 0.111 | atp6v0cb | -0.023 |
| si:ch211-243a20.4 | 0.111 | hbae3 | -0.023 |
| itgae.2 | 0.110 | stmn1b | -0.023 |
| adgrg3 | 0.109 | cspg5a | -0.022 |
| cmklr1 | 0.109 | icn | -0.022 |
| mpeg1.1 | 0.109 | dap1b | -0.022 |
| glipr1a | 0.107 | si:ch1073-429i10.3.1 | -0.022 |
| grap2b | 0.107 | actc1b | -0.021 |
| cxcr3.2 | 0.105 | fam168a | -0.021 |
| ccr9a | 0.104 | gng3 | -0.021 |
| LOC108180725 | 0.104 | hbae1.1 | -0.021 |
| cx32.2 | 0.103 | icn2 | -0.021 |
| CT027744.2 | 0.102 | krtt1c19e | -0.021 |
| ctss2.1 | 0.102 | krt5 | -0.020 |
| ptpn6 | 0.102 | si:ch211-195b11.3 | -0.020 |
| rnaset2l | 0.102 | anxa1a | -0.019 |
| gpr183a | 0.101 | ccni | -0.019 |
| havcr1 | 0.101 | cebpd | -0.019 |
| il1b | 0.101 | elavl3 | -0.019 |
| si:zfos-741a10.3 | 0.101 | hbbe1.1 | -0.019 |
| si:dkey-53k12.2 | 0.101 | ndrg2 | -0.019 |
| itgae.1 | 0.100 | nova2 | -0.019 |




