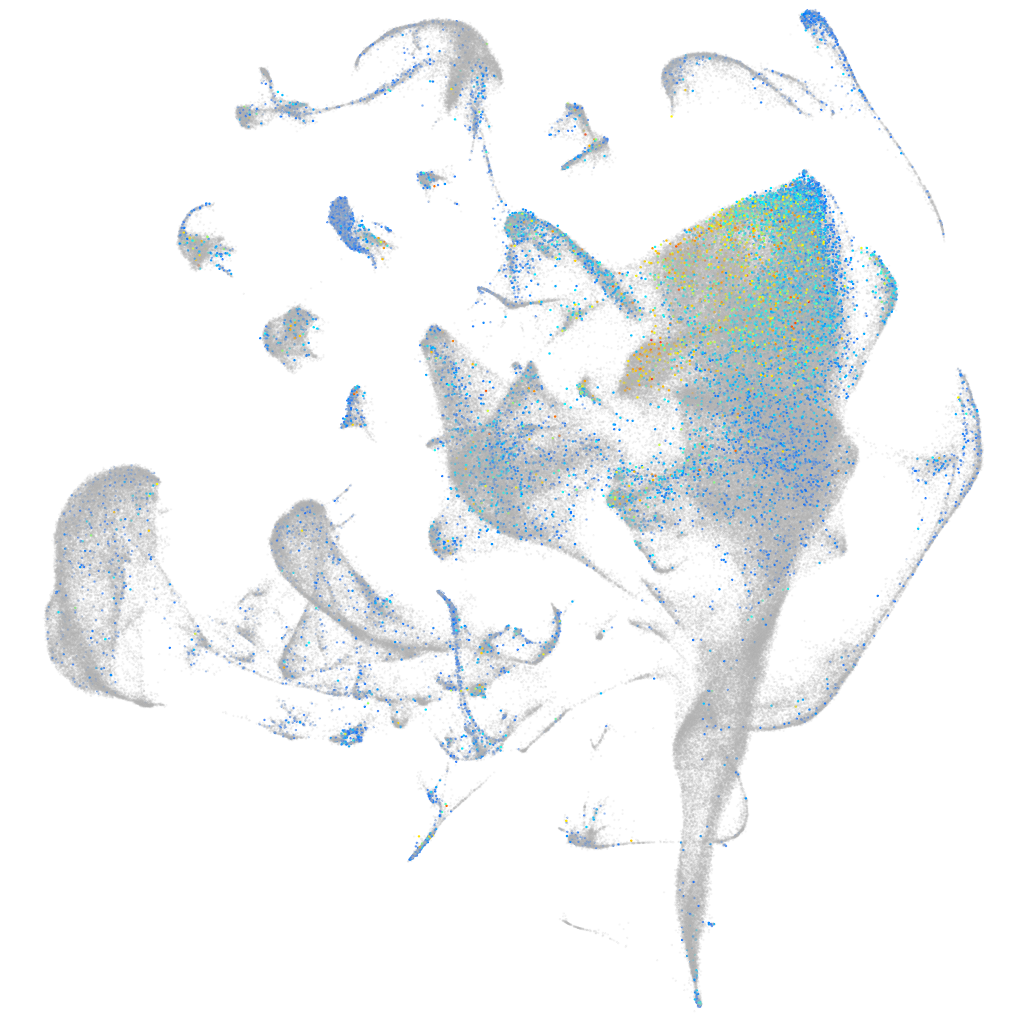thioesterase superfamily member 4
ZFIN 
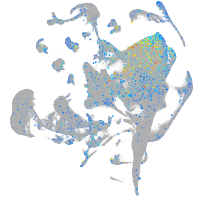
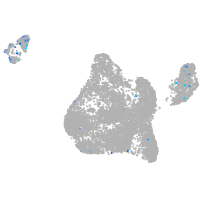
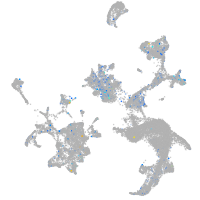
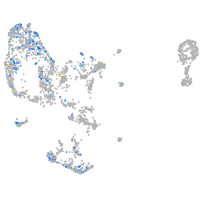
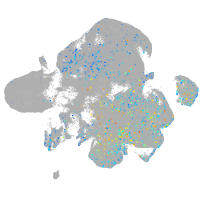
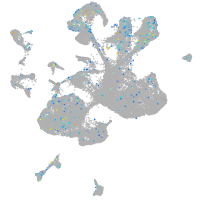
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ywhag2 | 0.127 | hspb1 | -0.059 |
| sncb | 0.125 | akap12b | -0.053 |
| gng3 | 0.124 | apoeb | -0.050 |
| zgc:65894 | 0.122 | id1 | -0.050 |
| atp5mc1 | 0.121 | si:ch211-152c2.3 | -0.048 |
| atp6v1e1b | 0.121 | stm | -0.048 |
| atp6v0cb | 0.120 | cfl1l | -0.046 |
| atp6v1g1 | 0.118 | pou5f3 | -0.046 |
| tpi1b | 0.118 | cx43.4 | -0.045 |
| gapdhs | 0.116 | eif4ebp3l | -0.045 |
| vamp2 | 0.116 | cast | -0.044 |
| stxbp1a | 0.116 | epcam | -0.044 |
| stmn2a | 0.114 | arf1 | -0.043 |
| rnasekb | 0.113 | polr3gla | -0.043 |
| ndufa4 | 0.112 | s100a1 | -0.043 |
| zgc:153426 | 0.112 | asb11 | -0.042 |
| mllt11 | 0.110 | bzw1b | -0.042 |
| syn2a | 0.110 | lye | -0.042 |
| stx1b | 0.109 | si:dkey-66i24.9 | -0.042 |
| ckbb | 0.108 | vox | -0.042 |
| snap25a | 0.108 | wu:fb18f06 | -0.042 |
| cnrip1a | 0.107 | zgc:56699 | -0.042 |
| si:ch73-119p20.1 | 0.106 | BX927258.1 | -0.041 |
| tuba1c | 0.106 | lig1 | -0.041 |
| cox7a2a | 0.105 | aldob | -0.040 |
| atpv0e2 | 0.103 | ckap4 | -0.040 |
| eno2 | 0.103 | sdc4 | -0.040 |
| map1aa | 0.103 | tbx16 | -0.040 |
| eno1a | 0.102 | ved | -0.040 |
| tmsb2 | 0.102 | ahnak | -0.039 |
| stmn1b | 0.100 | cyt1l | -0.039 |
| atp6ap2 | 0.099 | si:ch1073-80i24.3 | -0.039 |
| calm1a | 0.099 | anxa1a | -0.038 |
| calm2a | 0.098 | anxa1c | -0.038 |
| elavl4 | 0.097 | apela | -0.038 |