THAP domain containing 11
ZFIN 
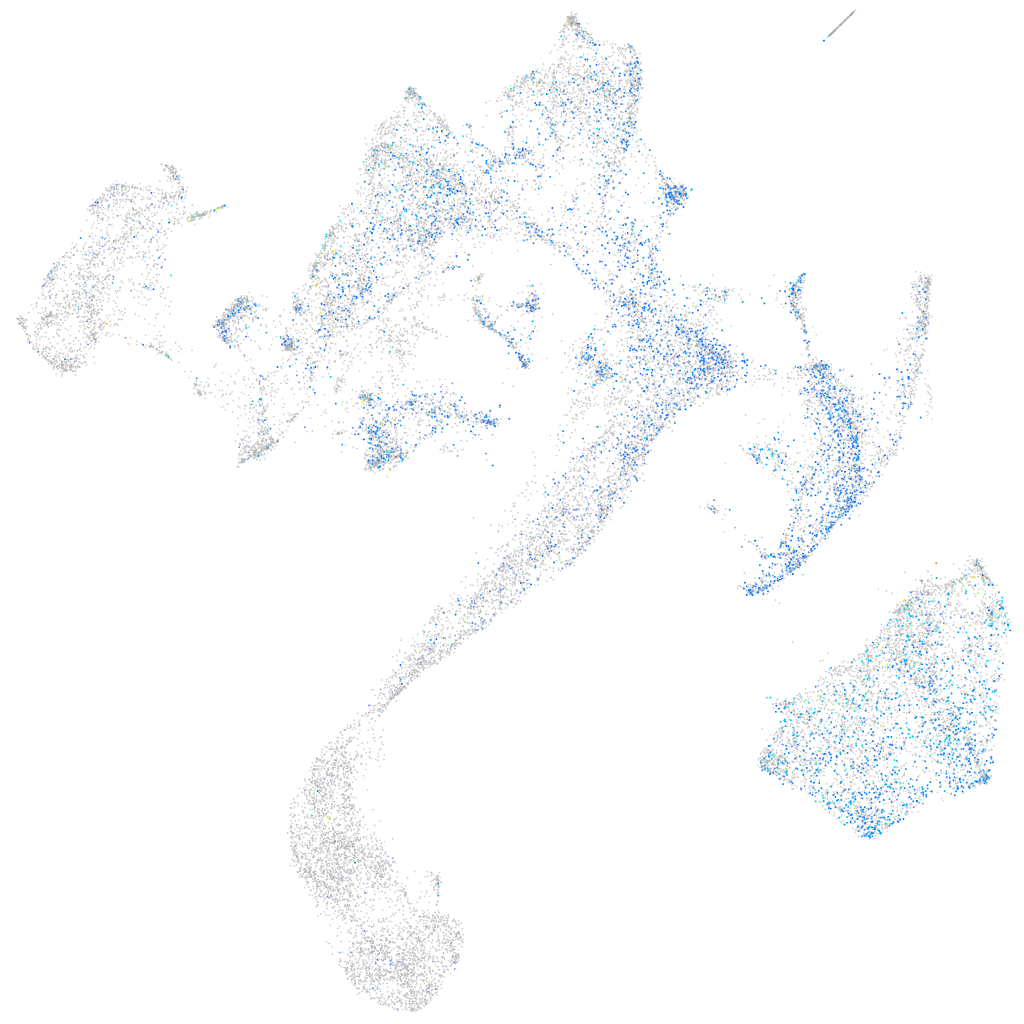
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| anp32b | 0.229 | actc1b | -0.211 |
| hmgb2a | 0.227 | ttn.2 | -0.190 |
| hmgb2b | 0.222 | aldoab | -0.189 |
| hnrnpabb | 0.221 | ckmb | -0.189 |
| hmga1a | 0.219 | atp2a1 | -0.188 |
| khdrbs1a | 0.219 | ckma | -0.188 |
| cbx3a | 0.217 | ak1 | -0.187 |
| seta | 0.217 | ttn.1 | -0.186 |
| hnrnpa0b | 0.216 | tnnc2 | -0.183 |
| si:ch211-222l21.1 | 0.215 | neb | -0.179 |
| ptmab | 0.215 | tmem38a | -0.179 |
| hnrnpaba | 0.213 | acta1b | -0.176 |
| anp32a | 0.213 | mybphb | -0.173 |
| tuba8l4 | 0.211 | mylpfa | -0.172 |
| h2afvb | 0.211 | ldb3a | -0.171 |
| si:ch73-1a9.3 | 0.210 | tpma | -0.171 |
| cirbpa | 0.209 | pabpc4 | -0.170 |
| sumo3a | 0.209 | ldb3b | -0.170 |
| cx43.4 | 0.209 | srl | -0.168 |
| si:ch211-288g17.3 | 0.209 | actn3b | -0.168 |
| srsf3b | 0.209 | desma | -0.167 |
| si:ch73-281n10.2 | 0.204 | CABZ01078594.1 | -0.167 |
| tubb2b | 0.202 | actn3a | -0.167 |
| hmgn7 | 0.202 | si:ch73-367p23.2 | -0.166 |
| ppm1g | 0.201 | gapdh | -0.165 |
| cirbpb | 0.201 | cav3 | -0.165 |
| hnrnpa1b | 0.200 | tnnt3a | -0.164 |
| setb | 0.200 | nme2b.2 | -0.163 |
| syncrip | 0.199 | acta1a | -0.162 |
| rbm8a | 0.198 | mylz3 | -0.161 |
| h3f3d | 0.198 | myom1a | -0.161 |
| nucks1a | 0.197 | myl1 | -0.161 |
| mki67 | 0.195 | eno1a | -0.161 |
| sumo3b | 0.194 | si:ch211-266g18.10 | -0.160 |
| marcksb | 0.194 | smyd1a | -0.159 |