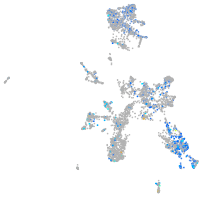
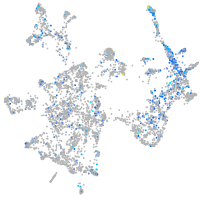
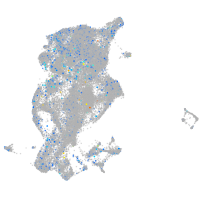
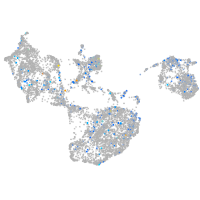
transferrin receptor 1b
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| bco2b | 0.190 | col1a1a | -0.049 |
| slc47a2.2 | 0.189 | bambia | -0.045 |
| CR376751.1 | 0.187 | bmp2b | -0.041 |
| si:dkey-18j18.3 | 0.147 | msx1b | -0.041 |
| BX005450.1 | 0.135 | ephb3 | -0.040 |
| CR396583.1 | 0.135 | col14a1a | -0.040 |
| inpp5jb | 0.130 | LOC101882251 | -0.040 |
| syngap1b | 0.130 | frem1a | -0.040 |
| SLCO3A1 (1 of many) | 0.127 | itih5 | -0.039 |
| arhgef33 | 0.122 | and1 | -0.038 |
| CR788254.2 | 0.119 | ecm2 | -0.037 |
| il1rapl2 | 0.118 | fras1 | -0.037 |
| buc | 0.118 | sp8a | -0.037 |
| CABZ01085140.1 | 0.117 | grip1 | -0.037 |
| CU207301.3 | 0.116 | nid1b | -0.036 |
| dlgap2a | 0.116 | mdka | -0.036 |
| LOC101882112 | 0.115 | lamb1a | -0.036 |
| zdhhc11 | 0.113 | sp9 | -0.036 |
| BX530017.2 | 0.113 | pls3 | -0.036 |
| si:dkey-57h18.1 | 0.112 | sp6 | -0.035 |
| or132-5 | 0.111 | egfl6 | -0.035 |
| CU462913.1 | 0.110 | fstl1a | -0.035 |
| MSANTD2 | 0.110 | rn7sk | -0.035 |
| wdr17 | 0.109 | frem2a | -0.034 |
| plcb4 | 0.108 | vox | -0.034 |
| mrap2b | 0.107 | hmcn1 | -0.034 |
| CT033796.1 | 0.106 | myofl | -0.034 |
| LOC103909923 | 0.106 | dlx2a | -0.034 |
| odf3l2a | 0.104 | lamc1 | -0.033 |
| CABZ01068273.1 | 0.103 | tmsb4x | -0.033 |
| lcp2a | 0.103 | sec61b | -0.033 |
| STX3 | 0.102 | NC-002333.4 | -0.033 |
| agxta | 0.101 | CABZ01072614.1 | -0.033 |
| faimb | 0.098 | ogna | -0.033 |
| grin2bb | 0.098 | CU459056.1 | -0.033 |