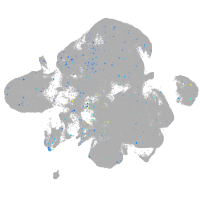
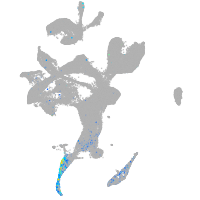
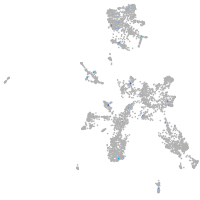
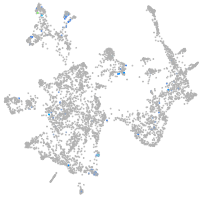
transferrin receptor 1a
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cox6a2 | 0.303 | si:ch73-1a9.3 | -0.216 |
| cox5b2 | 0.283 | ptmab | -0.209 |
| mt-nd2 | 0.283 | hmgn6 | -0.208 |
| mt-atp6 | 0.271 | h3f3d | -0.206 |
| si:ch211-139a5.9 | 0.268 | hnrnpaba | -0.197 |
| glud1b | 0.267 | si:ch73-281n10.2 | -0.196 |
| sod2 | 0.265 | si:ch211-222l21.1 | -0.196 |
| nnt | 0.262 | cirbpb | -0.182 |
| mt-nd4 | 0.262 | marcksb | -0.177 |
| chchd10 | 0.262 | hmga1a | -0.168 |
| sod1 | 0.257 | hmgb2a | -0.167 |
| mt-nd1 | 0.252 | hspb1 | -0.166 |
| si:dkey-16p21.8 | 0.251 | h2afvb | -0.166 |
| mt-co1 | 0.244 | nucks1a | -0.165 |
| crip1 | 0.240 | marcksl1b | -0.165 |
| COX3 | 0.240 | id3 | -0.163 |
| mtbl | 0.239 | hmgn2 | -0.160 |
| pck2 | 0.239 | hmgb3a | -0.158 |
| mt-co2 | 0.235 | anp32a | -0.157 |
| mt-nd3 | 0.233 | seta | -0.156 |
| epdl2 | 0.232 | hmgb1b | -0.155 |
| zgc:85777 | 0.231 | hmgb2b | -0.154 |
| ndufa11 | 0.231 | hnrnpabb | -0.153 |
| hadh | 0.229 | eef1a1l1 | -0.152 |
| alpl | 0.228 | hmgn7 | -0.150 |
| mt-cyb | 0.227 | hdac1 | -0.149 |
| aldh6a1 | 0.226 | setb | -0.147 |
| atp5f1c | 0.225 | apoeb | -0.147 |
| sdha | 0.225 | msna | -0.147 |
| galnt14 | 0.225 | wu:fb97g03 | -0.147 |
| suclg1 | 0.224 | h2afva | -0.145 |
| atp1a1a.4 | 0.223 | cx43.4 | -0.144 |
| cox7a2a | 0.222 | syncrip | -0.144 |
| cdh17 | 0.222 | apoc1 | -0.144 |
| mdh1aa | 0.222 | tubb2b | -0.142 |