transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma)
ZFIN 
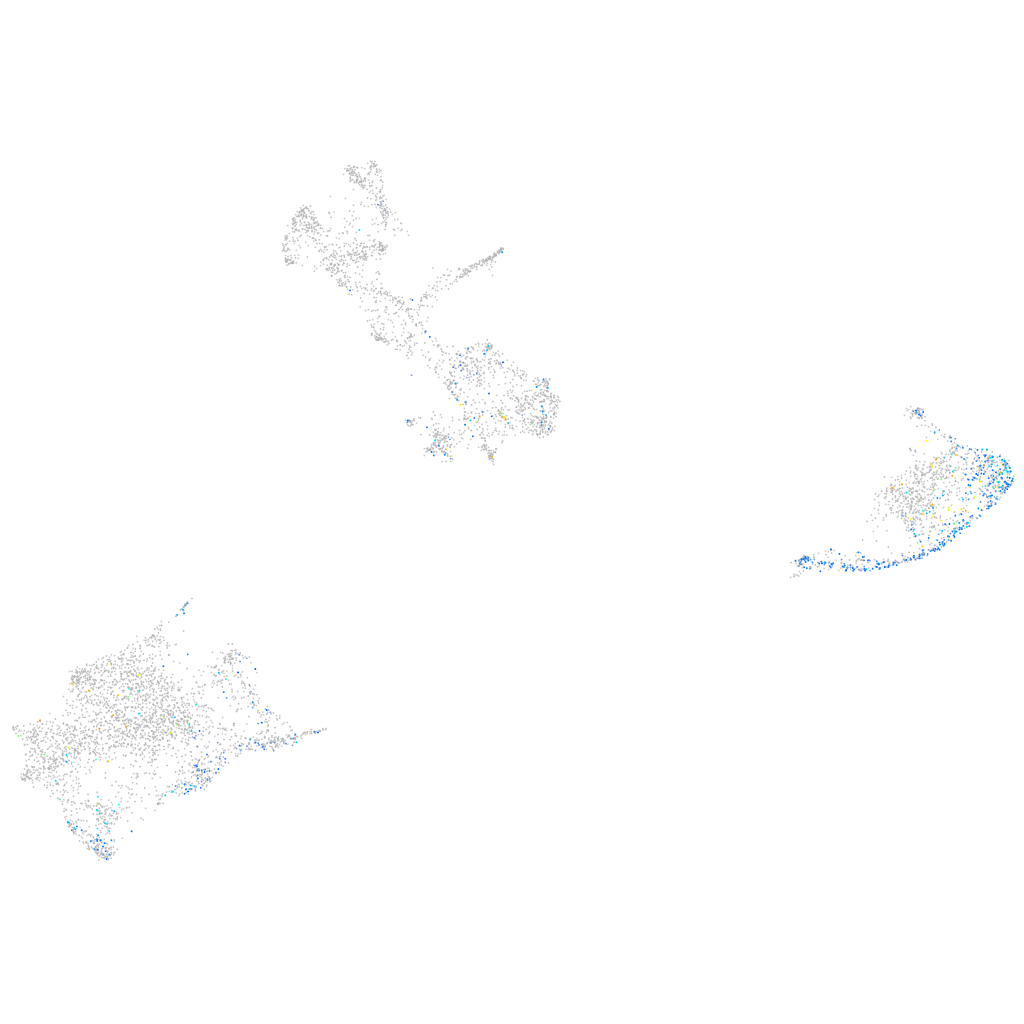
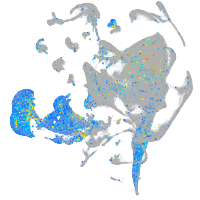
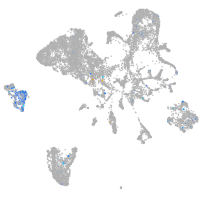
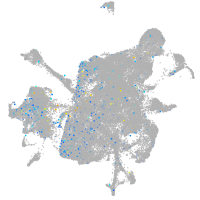
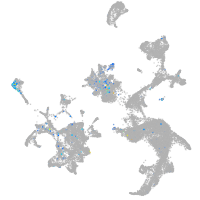
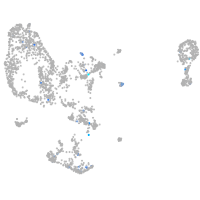
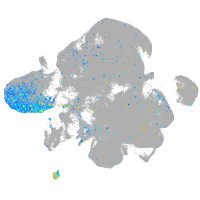
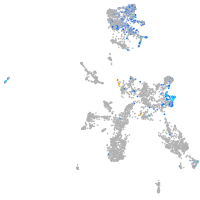
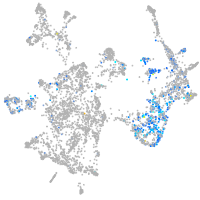
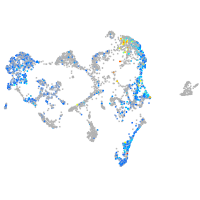
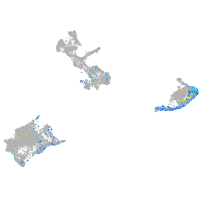
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| kita | 0.306 | paics | -0.157 |
| SPAG9 | 0.291 | uraha | -0.125 |
| oca2 | 0.266 | CABZ01021592.1 | -0.124 |
| pmela | 0.260 | mdh1aa | -0.124 |
| tfap2e | 0.257 | defbl1 | -0.116 |
| tyrp1b | 0.255 | apoda.1 | -0.112 |
| slc39a10 | 0.250 | impdh1b | -0.109 |
| tyrp1a | 0.248 | si:dkey-251i10.2 | -0.104 |
| dct | 0.246 | atic | -0.101 |
| si:ch73-389b16.1 | 0.244 | si:ch211-256m1.8 | -0.100 |
| lamp1a | 0.242 | gpnmb | -0.100 |
| slc24a5 | 0.239 | si:dkey-197i20.6 | -0.099 |
| zgc:91968 | 0.239 | lypc | -0.097 |
| mchr2 | 0.236 | tmem130 | -0.095 |
| tyr | 0.231 | phyhd1 | -0.095 |
| prkar1b | 0.230 | unm-sa821 | -0.092 |
| mtbl | 0.229 | si:ch73-89b15.3 | -0.091 |
| kcnj13 | 0.225 | aqp1a.1 | -0.090 |
| mitfa | 0.222 | glulb | -0.090 |
| hsd20b2 | 0.220 | akap12a | -0.090 |
| mlpha | 0.218 | slc2a15a | -0.088 |
| aadac | 0.212 | alx4a | -0.088 |
| gpr61 | 0.211 | alx4b | -0.086 |
| tfap2a | 0.209 | sytl2b | -0.084 |
| slc22a2 | 0.205 | ponzr1 | -0.082 |
| pknox1.2 | 0.201 | alx1 | -0.082 |
| slc24a4a | 0.201 | krt18b | -0.082 |
| slc45a2 | 0.194 | s100v2 | -0.081 |
| pah | 0.194 | pltp | -0.081 |
| slc7a5 | 0.193 | cdh11 | -0.079 |
| anxa1a | 0.191 | cx43 | -0.075 |
| gstt1a | 0.191 | prps1a | -0.075 |
| megf10 | 0.190 | slc25a36a | -0.075 |
| si:ch211-195b13.1 | 0.187 | sort1a | -0.075 |
| slc3a2a | 0.186 | tcirg1a | -0.074 |