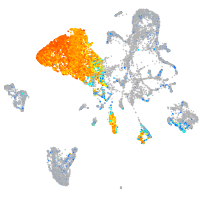
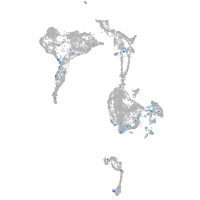
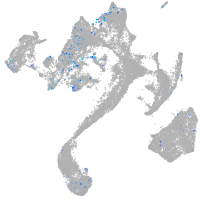
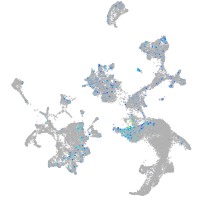
transferrin-a
ZFIN 
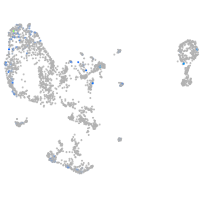
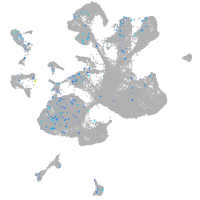
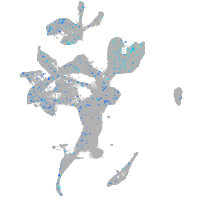
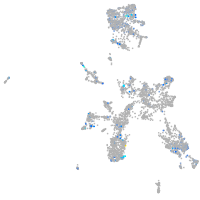
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| apoa2 | 0.243 | qdpra | -0.082 |
| apoa1b | 0.241 | prdx1 | -0.079 |
| prss59.2 | 0.195 | hsp90ab1 | -0.062 |
| c3a.5 | 0.167 | psmb7 | -0.062 |
| fmn1 | 0.148 | canx | -0.061 |
| ela3l | 0.143 | ptges3b | -0.061 |
| CELA1 (1 of many) | 0.142 | akr1b1 | -0.060 |
| FO704758.1 | 0.141 | abracl | -0.059 |
| masp2 | 0.138 | pts | -0.058 |
| prss1 | 0.138 | impdh1b | -0.058 |
| CABZ01072487.1 | 0.137 | rbp4l | -0.057 |
| etnppl | 0.136 | pcbd1 | -0.057 |
| a2ml | 0.136 | CABZ01032488.1 | -0.056 |
| ebi3 | 0.131 | dad1 | -0.055 |
| pygl | 0.128 | cst14b.1 | -0.055 |
| BX936364.1 | 0.128 | lman2 | -0.054 |
| ctrl | 0.126 | hmga1a | -0.052 |
| ctrb1 | 0.122 | abcf1 | -0.052 |
| hpn | 0.119 | rplp2l | -0.051 |
| ela2l | 0.118 | xbp1 | -0.050 |
| shbg | 0.116 | hnrnpabb | -0.050 |
| fabp10a | 0.116 | cfl1 | -0.050 |
| nipsnap1 | 0.115 | sub1a | -0.049 |
| ela2 | 0.115 | atp6v0ca | -0.049 |
| ambp | 0.114 | hspe1 | -0.049 |
| hamp | 0.113 | ssr4 | -0.049 |
| apoc1 | 0.111 | psmb6 | -0.048 |
| FO904898.4 | 0.111 | pdia3 | -0.048 |
| AL954146.1 | 0.110 | mibp | -0.048 |
| crp2 | 0.107 | eif5a2 | -0.048 |
| pvalb2 | 0.107 | slc37a2 | -0.048 |
| LOC110437989 | 0.106 | eif2s2 | -0.048 |
| gc | 0.105 | rpl13 | -0.048 |
| XLOC-008303 | 0.105 | rpl5b | -0.048 |
| krt17 | 0.105 | ost4 | -0.047 |