terminal nucleotidyltransferase 5Ab
ZFIN 
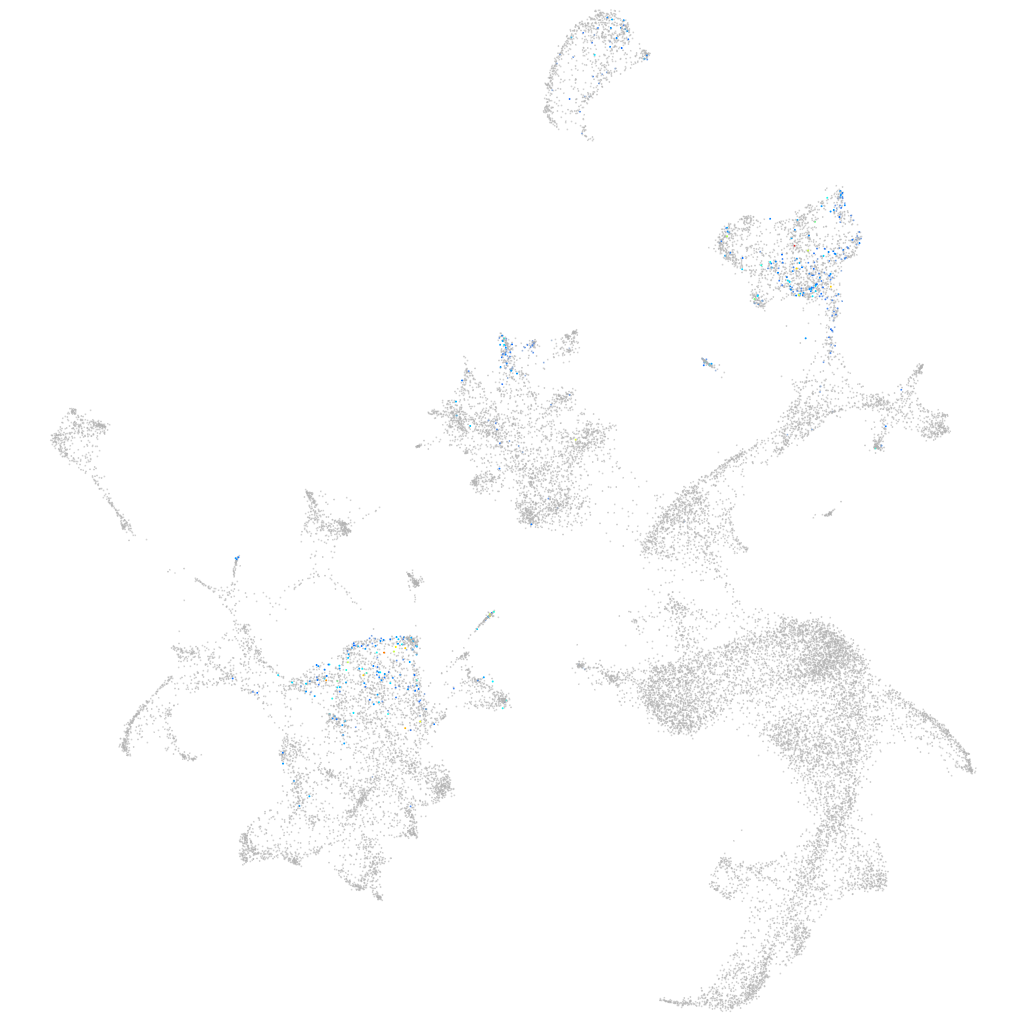
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme 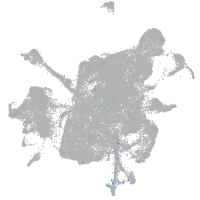 |
hematopoietic / vasculature 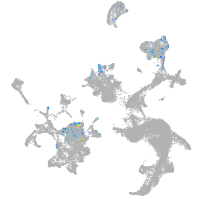 |
pronephros 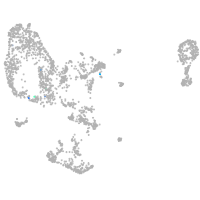 |
neural |
spinal cord / glia |
eye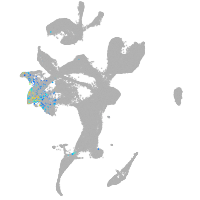 |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mrc1b | 0.175 | hbae1.1 | -0.109 |
| gnpda1 | 0.165 | hbae1.3 | -0.103 |
| stab2 | 0.157 | hbbe2 | -0.102 |
| tfec | 0.155 | hbae3 | -0.101 |
| zgc:158343 | 0.155 | hbbe1.3 | -0.101 |
| mafbb | 0.150 | hbbe1.1 | -0.099 |
| cotl1 | 0.149 | cahz | -0.096 |
| zgc:110239 | 0.146 | hemgn | -0.092 |
| mrc1a | 0.146 | hbbe1.2 | -0.091 |
| lgmn | 0.144 | alas2 | -0.087 |
| dab2 | 0.142 | fth1a | -0.086 |
| lamp2 | 0.140 | blvrb | -0.085 |
| tspan36 | 0.139 | slc4a1a | -0.083 |
| naga | 0.136 | nt5c2l1 | -0.081 |
| cndp2 | 0.136 | si:ch211-250g4.3 | -0.080 |
| ctsh | 0.135 | epb41b | -0.079 |
| asah1b | 0.134 | zgc:163057 | -0.077 |
| ctsba | 0.134 | zgc:56095 | -0.077 |
| zgc:162730 | 0.133 | tspo | -0.076 |
| hexb | 0.133 | creg1 | -0.074 |
| si:dkey-28n18.9 | 0.132 | nmt1b | -0.072 |
| slc3a2a | 0.131 | si:ch211-207c6.2 | -0.070 |
| snx1a | 0.131 | prdx2 | -0.068 |
| zgc:110591 | 0.131 | plac8l1 | -0.067 |
| scpep1 | 0.131 | tmod4 | -0.067 |
| cebpb | 0.131 | znfl2a | -0.063 |
| cd63 | 0.131 | tfr1a | -0.062 |
| zfp36l1a | 0.130 | sptb | -0.062 |
| mafba | 0.130 | rfesd | -0.061 |
| mfap4 | 0.130 | uros | -0.061 |
| lyve1b | 0.128 | hdr | -0.061 |
| glula | 0.128 | si:ch211-227m13.1 | -0.059 |
| cst14b.1 | 0.127 | hbae5 | -0.058 |
| si:zfos-1069f5.1 | 0.127 | hbbe3 | -0.057 |
| atp6v0ca | 0.127 | TCIM (1 of many) | -0.057 |