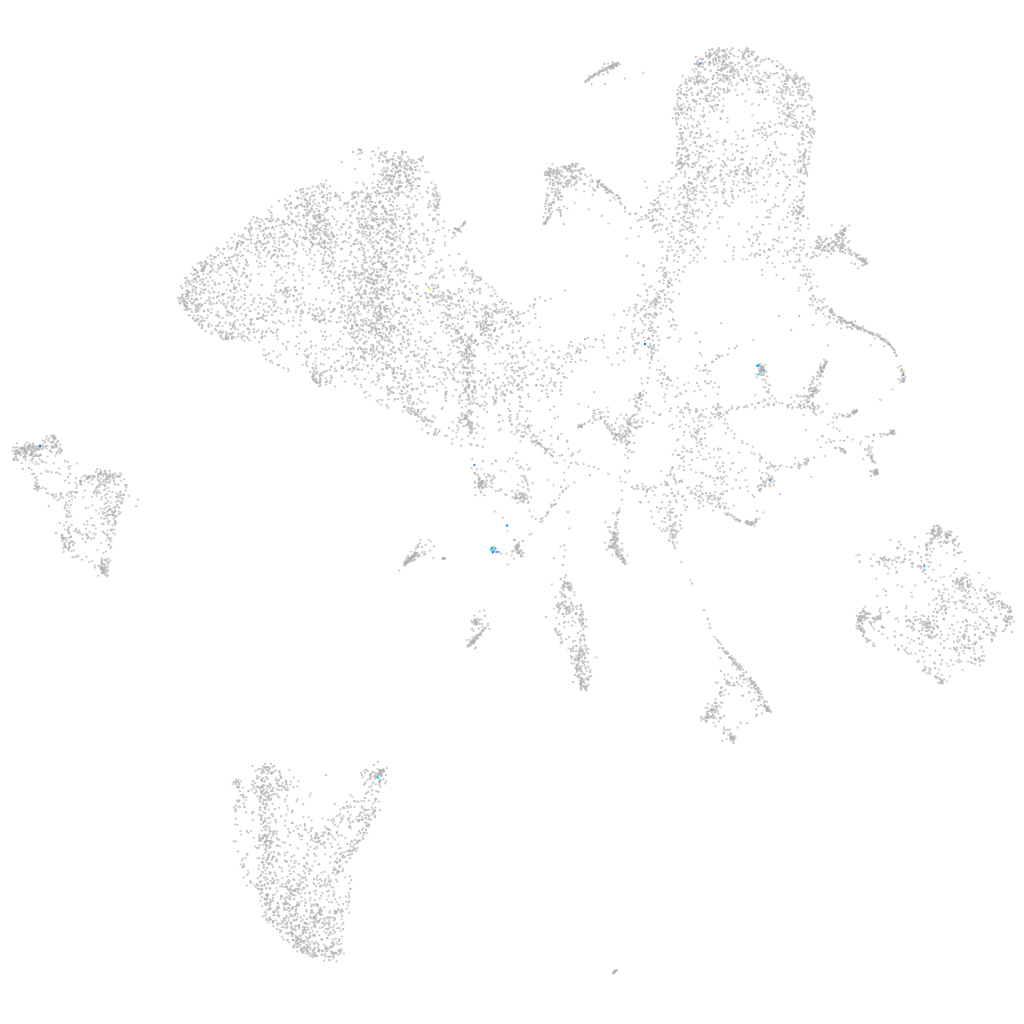
tektin 1
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural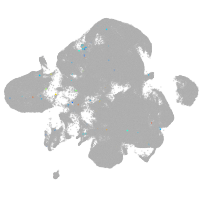 |
spinal cord / glia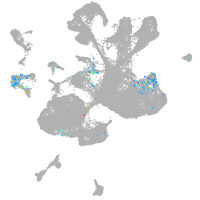 |
eye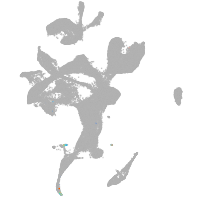 |
taste / olfactory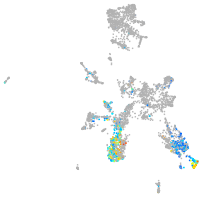 |
otic / lateral line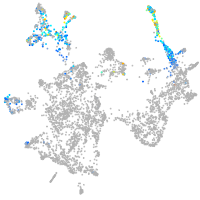 |
epidermis |
periderm |
fin |
ionocytes / mucous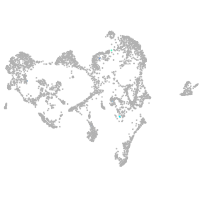 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cdc25d | 0.557 | gatm | -0.039 |
| spata18 | 0.491 | gamt | -0.037 |
| zgc:195356 | 0.481 | fabp3 | -0.035 |
| LOC103911624 | 0.440 | cdo1 | -0.033 |
| si:ch211-198b3.4 | 0.415 | dap | -0.033 |
| iqcg | 0.414 | gapdh | -0.032 |
| efhb | 0.375 | ahcy | -0.032 |
| si:dkey-148f10.4 | 0.368 | apobb.1 | -0.032 |
| si:dkey-76p14.2 | 0.365 | hdlbpa | -0.032 |
| crocc2 | 0.356 | slc27a2a | -0.031 |
| dnah5l | 0.350 | apoa1b | -0.031 |
| si:ch211-155m12.5 | 0.345 | nupr1b | -0.031 |
| pacrg | 0.344 | afp4 | -0.031 |
| si:ch211-130h14.4 | 0.340 | apoa4b.1 | -0.031 |
| casc1 | 0.332 | abat | -0.030 |
| tex36 | 0.326 | apoc2 | -0.030 |
| tbata | 0.320 | cx32.3 | -0.030 |
| rsph14 | 0.316 | rgn | -0.029 |
| capsla | 0.311 | gnmt | -0.029 |
| gmnc | 0.310 | cox7a1 | -0.029 |
| theg | 0.307 | fbp1b | -0.029 |
| efhc1 | 0.306 | gstm.1 | -0.029 |
| CFAP77 | 0.305 | pck2 | -0.029 |
| dnaaf3 | 0.300 | cx28.9 | -0.029 |
| nme8 | 0.299 | bhmt | -0.029 |
| spaca9 | 0.290 | hspd1 | -0.028 |
| cfap45 | 0.283 | si:ch211-284e20.8 | -0.028 |
| XLOC-004950 | 0.279 | c8g | -0.028 |
| LOC110438396 | 0.279 | msrb2 | -0.028 |
| LOC103911030 | 0.277 | rpl6 | -0.028 |
| CAPN14 | 0.277 | serpinf2b | -0.028 |
| nme5 | 0.274 | glud1b | -0.028 |
| ccdc65 | 0.273 | zgc:112265 | -0.027 |
| LOC101885015 | 0.270 | tfa | -0.027 |
| dydc2 | 0.268 | gstt1a | -0.027 |