tyrosyl-DNA phosphodiesterase 1
ZFIN 
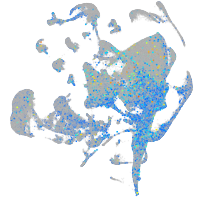
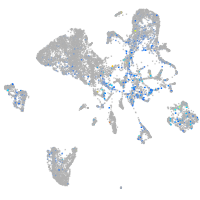
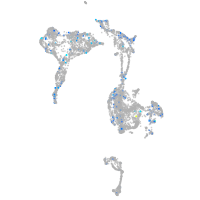
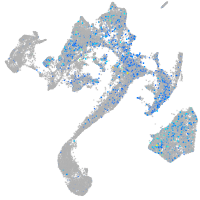
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pcna | 0.202 | rtn1a | -0.098 |
| dut | 0.197 | ckbb | -0.098 |
| rrm1 | 0.189 | elavl3 | -0.092 |
| stmn1a | 0.181 | myt1b | -0.088 |
| rrm2 | 0.181 | rnasekb | -0.081 |
| lig1 | 0.178 | gpm6ab | -0.081 |
| rpa2 | 0.174 | CR383676.1 | -0.081 |
| chaf1a | 0.173 | si:dkeyp-75h12.5 | -0.074 |
| fen1 | 0.173 | atp6v0cb | -0.073 |
| dek | 0.169 | gpm6aa | -0.073 |
| mibp | 0.168 | stmn1b | -0.072 |
| mcm7 | 0.168 | syt2a | -0.072 |
| banf1 | 0.167 | calm1a | -0.070 |
| anp32b | 0.167 | marcksl1b | -0.070 |
| nutf2l | 0.165 | gapdhs | -0.070 |
| cks1b | 0.165 | vamp2 | -0.069 |
| rpa3 | 0.163 | myt1a | -0.069 |
| zgc:110540 | 0.163 | atp6v1e1b | -0.068 |
| nasp | 0.162 | stx1b | -0.068 |
| rnaseh2a | 0.162 | stxbp1a | -0.067 |
| ccna2 | 0.161 | dpysl2b | -0.067 |
| dhfr | 0.158 | slc1a2b | -0.066 |
| CABZ01005379.1 | 0.156 | cotl1 | -0.066 |
| selenoh | 0.156 | dpysl3 | -0.066 |
| tuba8l4 | 0.156 | tmsb | -0.066 |
| ranbp1 | 0.153 | gng3 | -0.065 |
| hmgb2a | 0.153 | gnao1a | -0.065 |
| zgc:110216 | 0.152 | fez1 | -0.065 |
| mki67 | 0.152 | elavl4 | -0.064 |
| esco2 | 0.151 | sncb | -0.064 |
| lbr | 0.149 | ppdpfb | -0.064 |
| ssrp1a | 0.148 | pvalb1 | -0.063 |
| si:dkey-6i22.5 | 0.146 | cadm4 | -0.062 |
| prim2 | 0.146 | cplx2 | -0.062 |
| cdca5 | 0.146 | snap25a | -0.062 |