"TAF7 RNA polymerase II, TATA box binding protein (TBP)-associated factor"
ZFIN 
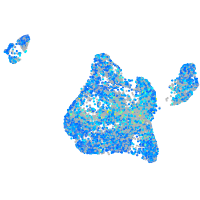
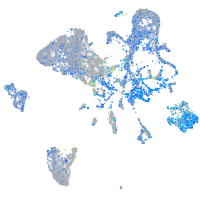
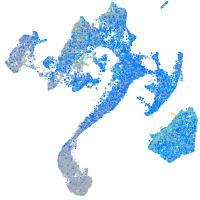
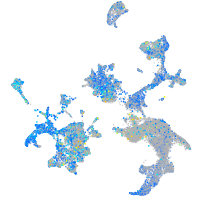
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hnrnpaba | 0.166 | atp1a1b | -0.130 |
| hnrnpa0l | 0.165 | fabp7a | -0.110 |
| khdrbs1a | 0.153 | ptn | -0.104 |
| h3f3d | 0.150 | slc1a2b | -0.096 |
| tubb2b | 0.148 | cx43 | -0.089 |
| hsp90ab1 | 0.145 | glula | -0.088 |
| hnrnpa0b | 0.143 | CU467822.1 | -0.087 |
| hnrnpabb | 0.143 | pvalb1 | -0.086 |
| sumo3a | 0.142 | slc4a4a | -0.085 |
| ptmab | 0.138 | si:ch211-66e2.5 | -0.084 |
| cirbpb | 0.136 | atp1b4 | -0.083 |
| hdac1 | 0.135 | si:ch211-251b21.1 | -0.081 |
| h3f3a | 0.133 | pvalb2 | -0.080 |
| ilf2 | 0.131 | actc1b | -0.080 |
| cct5 | 0.131 | qki2 | -0.080 |
| cirbpa | 0.131 | slc3a2a | -0.080 |
| hspa8 | 0.131 | mdkb | -0.079 |
| h2afvb | 0.126 | ppap2d | -0.076 |
| hnrnpa0a | 0.125 | efhd1 | -0.076 |
| smarce1 | 0.125 | apoa2 | -0.076 |
| syncrip | 0.125 | mt2 | -0.074 |
| cct2 | 0.125 | gpr37l1b | -0.074 |
| si:ch211-288g17.3 | 0.124 | cd63 | -0.074 |
| fkbp1aa | 0.123 | hepacama | -0.073 |
| hmgn6 | 0.123 | gpm6bb | -0.073 |
| si:ch1073-429i10.3.1 | 0.122 | apoa1b | -0.072 |
| si:ch211-51e12.7 | 0.122 | cox4i2 | -0.071 |
| cct3 | 0.122 | zgc:165461 | -0.071 |
| eif3ea | 0.121 | slc6a9 | -0.070 |
| slc25a5 | 0.121 | slc1a3b | -0.069 |
| snrpd2 | 0.120 | zgc:158463 | -0.068 |
| hmga1a | 0.119 | slc6a11b | -0.067 |
| si:ch211-222l21.1 | 0.119 | zgc:153704 | -0.067 |
| ran | 0.118 | cldn19 | -0.065 |
| oaz1b | 0.118 | smox | -0.064 |