"TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor"
ZFIN 
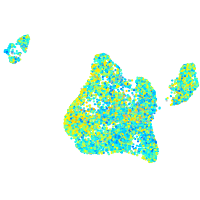
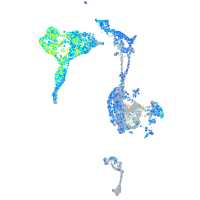
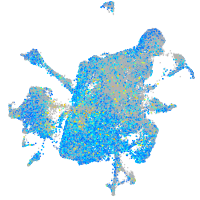
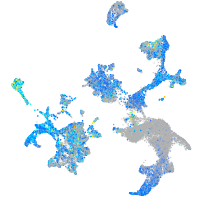
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hmga1a | 0.428 | si:ch211-139a5.9 | -0.347 |
| hmgb2b | 0.427 | eno3 | -0.318 |
| seta | 0.424 | atp1b1a | -0.316 |
| hnrnpabb | 0.414 | rpl37 | -0.310 |
| hnrnpaba | 0.410 | slc25a5 | -0.302 |
| ilf3b | 0.405 | atp5mc3b | -0.302 |
| hspb1 | 0.404 | ldhba | -0.299 |
| nucks1a | 0.402 | vdac3 | -0.282 |
| hnrnpa1a | 0.397 | mdh1aa | -0.279 |
| setb | 0.396 | atp5if1b | -0.276 |
| si:ch211-288g17.3 | 0.393 | atp5fa1 | -0.276 |
| cirbpa | 0.392 | rps10 | -0.271 |
| hnrnpa1b | 0.387 | dap1b | -0.267 |
| h2afvb | 0.387 | atp5f1b | -0.263 |
| snrpe | 0.386 | atp1a1a.4 | -0.262 |
| wu:fb97g03 | 0.385 | atp5mc1 | -0.259 |
| cx43.4 | 0.383 | cdh17 | -0.253 |
| snrpd1 | 0.379 | krt8 | -0.252 |
| snrpd2 | 0.377 | cdaa | -0.250 |
| ptges3b | 0.373 | romo1 | -0.249 |
| marcksb | 0.372 | si:dkey-16p21.8 | -0.246 |
| khdrbs1a | 0.372 | aldob | -0.245 |
| hnrnpa0b | 0.370 | zgc:114188 | -0.245 |
| syncrip | 0.368 | atp5l | -0.245 |
| anp32e | 0.368 | tpi1b | -0.243 |
| cbx3a | 0.368 | suclg1 | -0.243 |
| snu13b | 0.367 | rps17 | -0.241 |
| ilf2 | 0.366 | mt-co2 | -0.238 |
| apoc1 | 0.364 | atp5f1e | -0.238 |
| hdac1 | 0.363 | atp5pf | -0.238 |
| snrpf | 0.362 | aldh6a1 | -0.237 |
| hmgb2a | 0.360 | atp5meb | -0.237 |
| nop58 | 0.359 | selenow1 | -0.236 |
| hnrnpub | 0.357 | cox6a1 | -0.235 |
| top1l | 0.354 | cox7a1 | -0.232 |