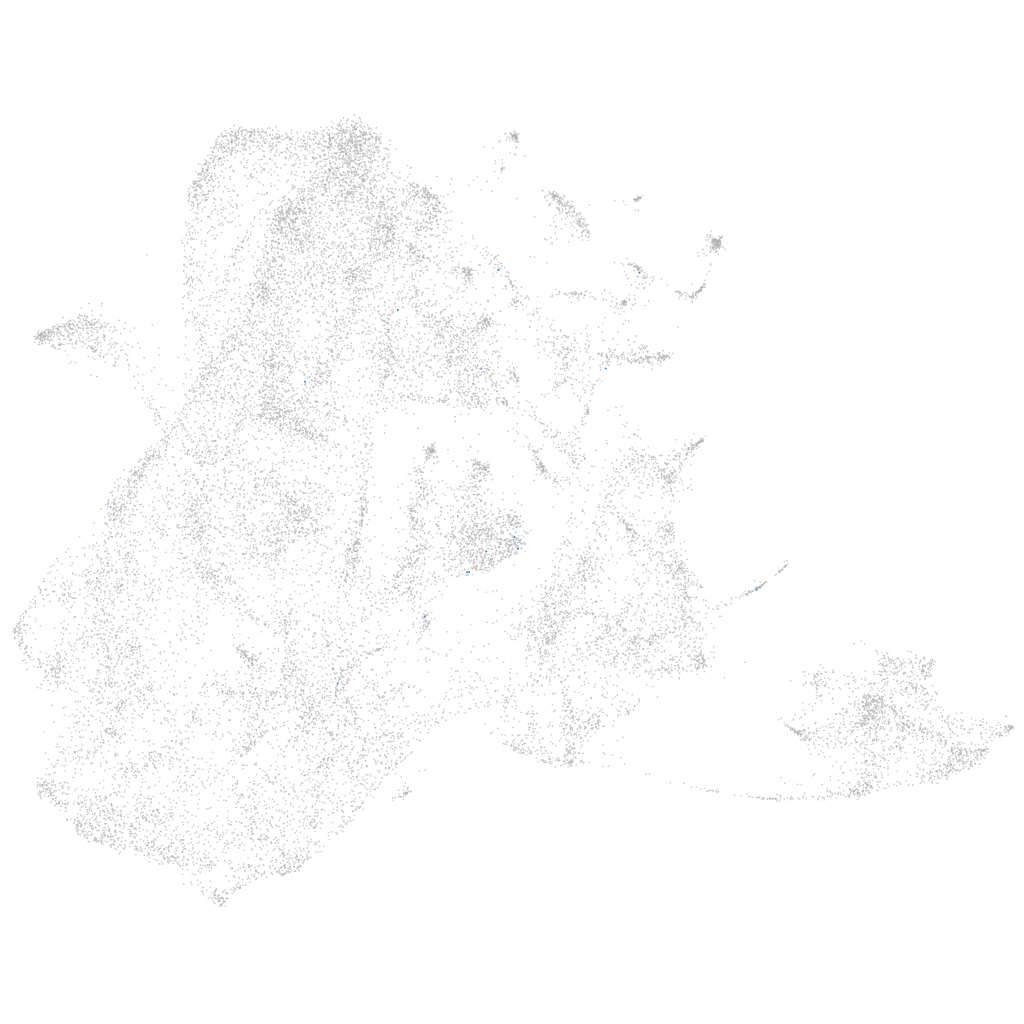
tachykinin precursor 3b
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 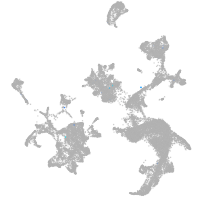 |
pronephros  |
neural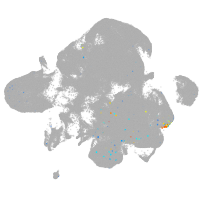 |
spinal cord / glia |
eye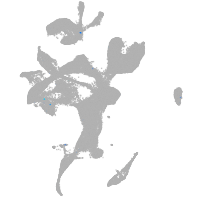 |
taste / olfactory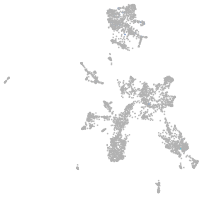 |
otic / lateral line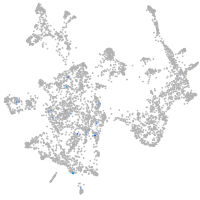 |
epidermis |
periderm |
fin |
ionocytes / mucous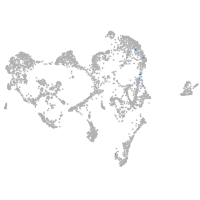 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:173915 | 0.233 | cfl1l | -0.052 |
| kiss2 | 0.228 | tmsb1 | -0.045 |
| prokr1b | 0.223 | pfn1 | -0.040 |
| LOC101886196 | 0.209 | epcam | -0.038 |
| CU207241.1 | 0.201 | zgc:153867 | -0.035 |
| LOC100537369 | 0.198 | actb2 | -0.035 |
| vipr1b | 0.168 | ccng1 | -0.033 |
| tacr2 | 0.166 | myl12.1 | -0.032 |
| htr1aa | 0.141 | arhgef5 | -0.031 |
| oprd1b | 0.138 | zgc:162730 | -0.030 |
| nmbr | 0.138 | myh9a | -0.030 |
| CR933734.2 | 0.134 | cldni | -0.030 |
| glrba | 0.133 | cldn1 | -0.029 |
| cldn2 | 0.128 | krt4 | -0.029 |
| CU928046.1 | 0.120 | s100a10b | -0.028 |
| abcd2 | 0.120 | tmsb4x | -0.027 |
| adora1b | 0.116 | sparc | -0.026 |
| CR753874.1 | 0.115 | flna | -0.026 |
| LOC103908739 | 0.115 | tagln2 | -0.026 |
| LO017848.1 | 0.115 | jupa | -0.025 |
| CABZ01113981.1 | 0.115 | krt8 | -0.025 |
| syt10 | 0.113 | ywhaz | -0.025 |
| si:dkey-166d12.2 | 0.113 | carhsp1 | -0.025 |
| nmur3 | 0.112 | pleca | -0.025 |
| CT476811.1 | 0.112 | pak2a | -0.025 |
| gabrb2 | 0.112 | sult2st1 | -0.025 |
| tnk2a | 0.111 | lgals3b | -0.024 |
| unm-sa808 | 0.109 | tpm3 | -0.024 |
| slc12a5b | 0.108 | capza1a | -0.023 |
| ecel1 | 0.107 | actb1 | -0.023 |
| si:ch211-188c16.1 | 0.106 | rplp2l | -0.023 |
| pou3f2a | 0.104 | cebpd | -0.023 |
| XLOC-043097 | 0.103 | zgc:171775 | -0.022 |
| zgc:153031 | 0.103 | spint1a | -0.022 |
| CR385067.1 | 0.103 | pxna | -0.022 |