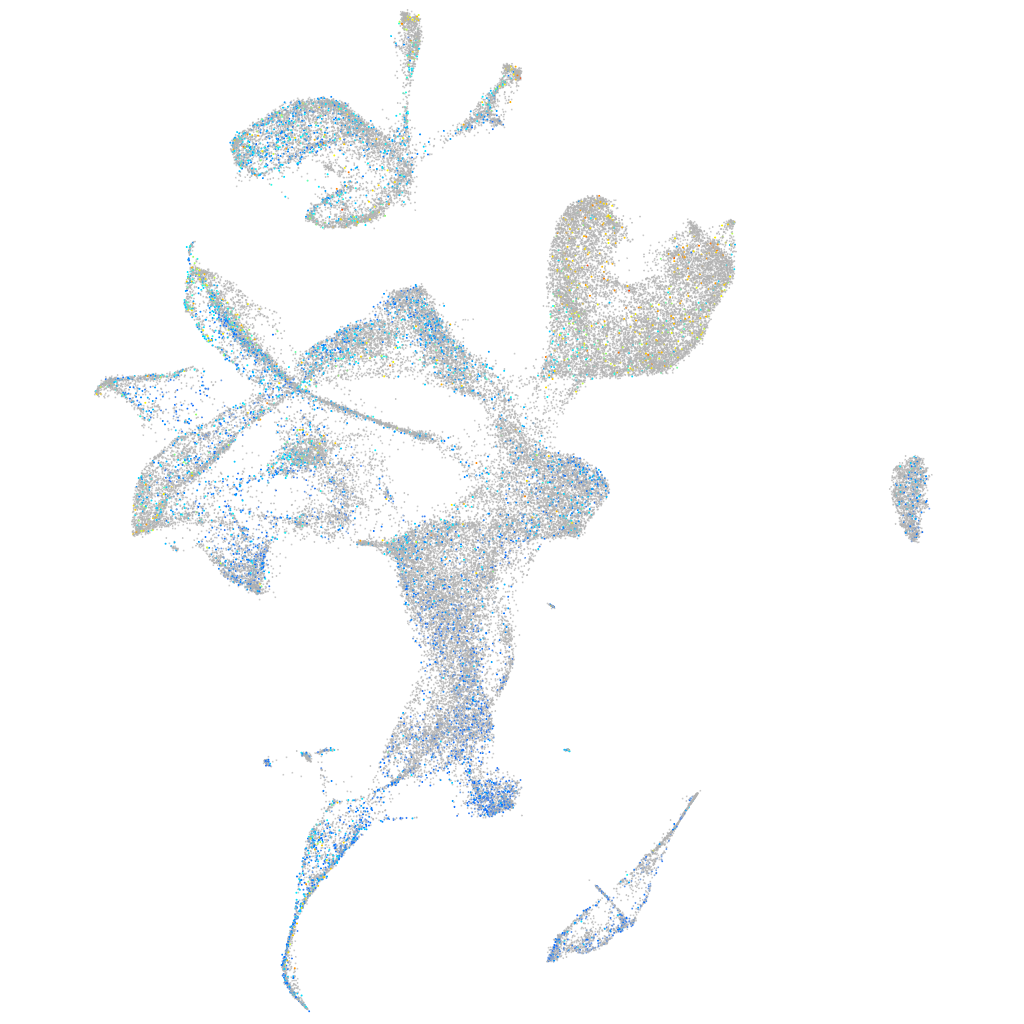
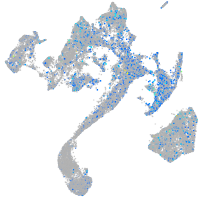
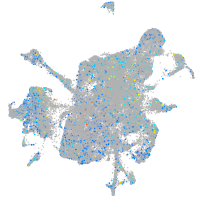
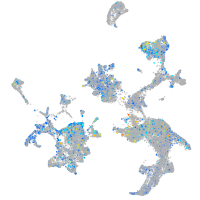
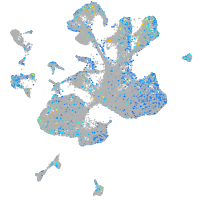
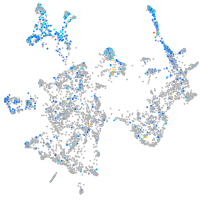
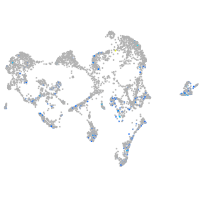
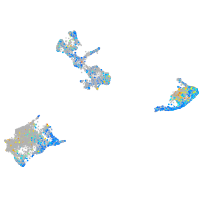
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gpr143 | 0.091 | hmgb2a | -0.071 |
| sncb | 0.090 | rrm1 | -0.058 |
| slc45a2 | 0.088 | hes2.2 | -0.054 |
| zgc:65894 | 0.085 | her15.1 | -0.053 |
| tspan36 | 0.085 | dla | -0.053 |
| eno2 | 0.085 | stmn1a | -0.052 |
| stmn2a | 0.083 | LOC798783 | -0.050 |
| dct | 0.083 | her4.2 | -0.049 |
| syngr1a | 0.082 | pcna | -0.049 |
| rtn1b | 0.082 | mki67 | -0.048 |
| gnb1a | 0.082 | hes6 | -0.048 |
| stxbp1a | 0.082 | si:ch73-1a9.3 | -0.048 |
| rab38 | 0.081 | si:ch211-222l21.1 | -0.048 |
| pmela | 0.080 | rrm2 | -0.048 |
| tspan10 | 0.079 | hmgn2 | -0.048 |
| ywhag2 | 0.078 | CABZ01005379.1 | -0.047 |
| pmelb | 0.077 | foxn4 | -0.047 |
| snap25a | 0.077 | hmga1a | -0.046 |
| atp6v1e1b | 0.077 | ccnd1 | -0.046 |
| tyrp1b | 0.077 | lig1 | -0.045 |
| gng3 | 0.076 | esco2 | -0.045 |
| vat1 | 0.076 | hmgb2b | -0.045 |
| tyrp1a | 0.076 | XLOC-003692 | -0.045 |
| mitfa | 0.075 | nasp | -0.045 |
| zgc:110591 | 0.075 | ccna2 | -0.045 |
| cplx2l | 0.075 | notch1a | -0.045 |
| qdpra | 0.075 | XLOC-042899 | -0.044 |
| oca2 | 0.074 | XLOC-003690 | -0.044 |
| bace2 | 0.074 | XLOC-003689 | -0.043 |
| zgc:153426 | 0.073 | her4.1 | -0.043 |
| rab32a | 0.073 | hist1h4l | -0.043 |
| hist2h2l | 0.073 | cdc25b | -0.043 |
| ywhaz | 0.073 | fbxo5 | -0.042 |
| slc24a5 | 0.073 | sox18 | -0.042 |
| FP085398.1 | 0.073 | slbp | -0.042 |