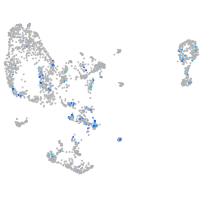
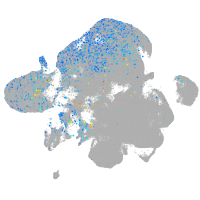
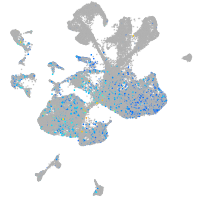
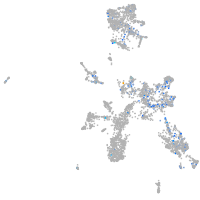
synaptophysin-like 2b
ZFIN 
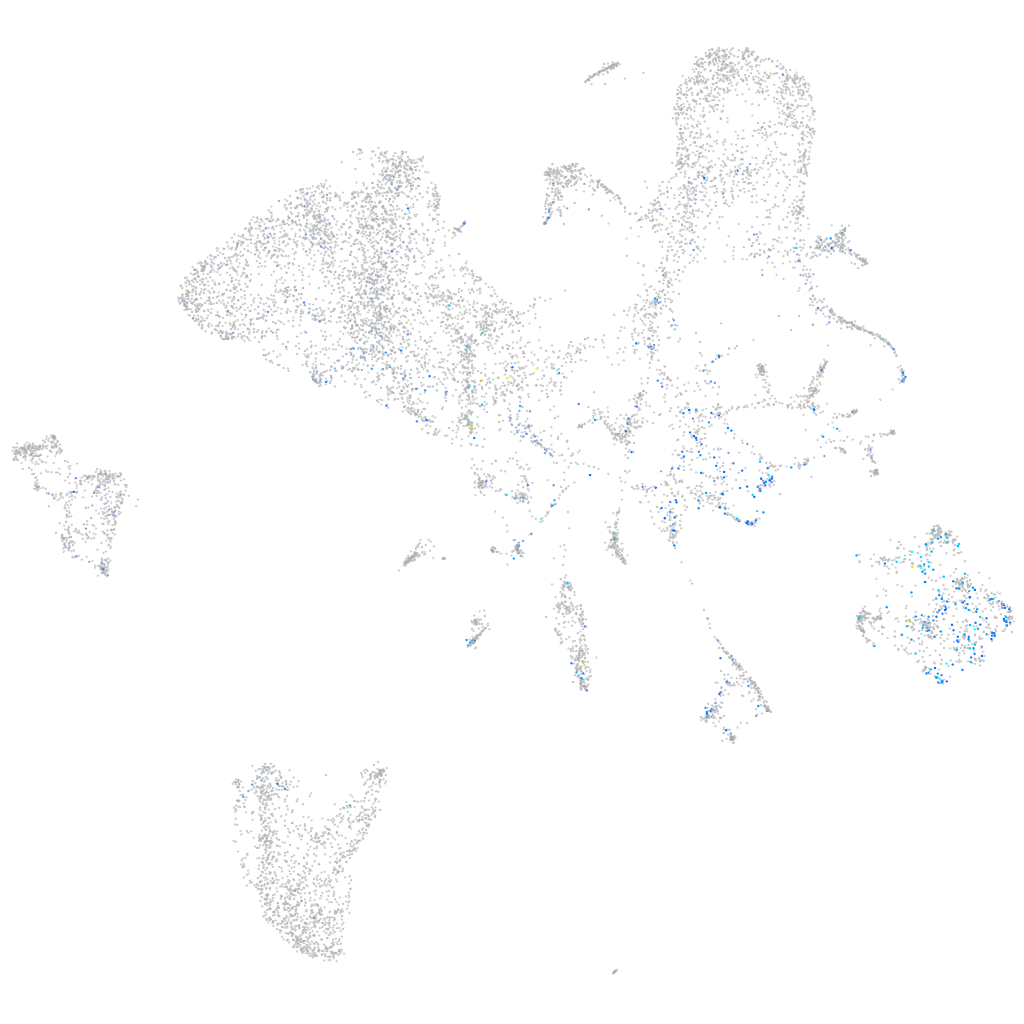
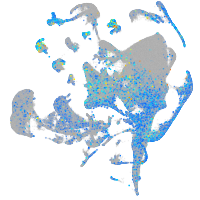
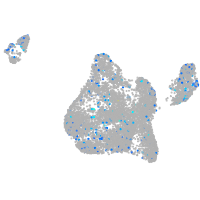
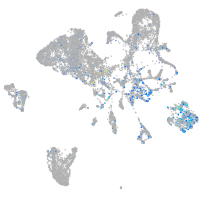
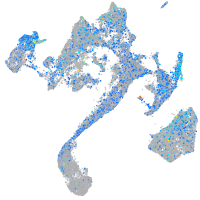
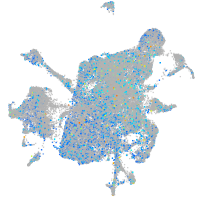
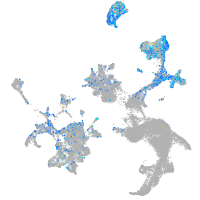
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hspb1 | 0.253 | rpl37 | -0.181 |
| akap12b | 0.251 | rps10 | -0.160 |
| cx43.4 | 0.247 | zgc:114188 | -0.160 |
| sp5l | 0.247 | nme2b.1 | -0.152 |
| sox17 | 0.242 | rps17 | -0.145 |
| foxd5 | 0.241 | gamt | -0.139 |
| si:ch211-152c2.3 | 0.239 | ahcy | -0.138 |
| marcksb | 0.235 | gapdh | -0.138 |
| sox32 | 0.230 | sod1 | -0.136 |
| flrt3 | 0.230 | atp5if1b | -0.129 |
| cdh6 | 0.228 | gatm | -0.127 |
| cxcr4a | 0.226 | eno3 | -0.125 |
| marcksl1b | 0.225 | zgc:158463 | -0.123 |
| ilf3b | 0.222 | dap | -0.120 |
| hnrnpub | 0.222 | suclg1 | -0.119 |
| sb:cb81 | 0.221 | gpx4a | -0.119 |
| stm | 0.219 | nupr1b | -0.119 |
| syncrip | 0.217 | lgals2b | -0.116 |
| hmga1a | 0.216 | fbp1b | -0.116 |
| nucks1a | 0.216 | glud1b | -0.115 |
| dhrs3b | 0.214 | zgc:92744 | -0.114 |
| smarca4a | 0.213 | mt-atp6 | -0.109 |
| snrnp70 | 0.213 | apoa4b.1 | -0.109 |
| qkia | 0.213 | eef1da | -0.108 |
| rbm4.3 | 0.212 | tpi1b | -0.107 |
| top1l | 0.212 | prdx2 | -0.107 |
| tp53inp2 | 0.211 | apoc2 | -0.106 |
| ca9 | 0.211 | mt-nd1 | -0.106 |
| safb | 0.210 | pnp4b | -0.106 |
| anp32b | 0.210 | cox6a1 | -0.105 |
| hnrnpa1b | 0.210 | cox7a1 | -0.105 |
| tbx16 | 0.209 | ckba | -0.104 |
| LOC100008030 | 0.209 | abat | -0.104 |
| smc1al | 0.209 | cx32.3 | -0.103 |
| ppm1g | 0.208 | srd5a2a | -0.102 |