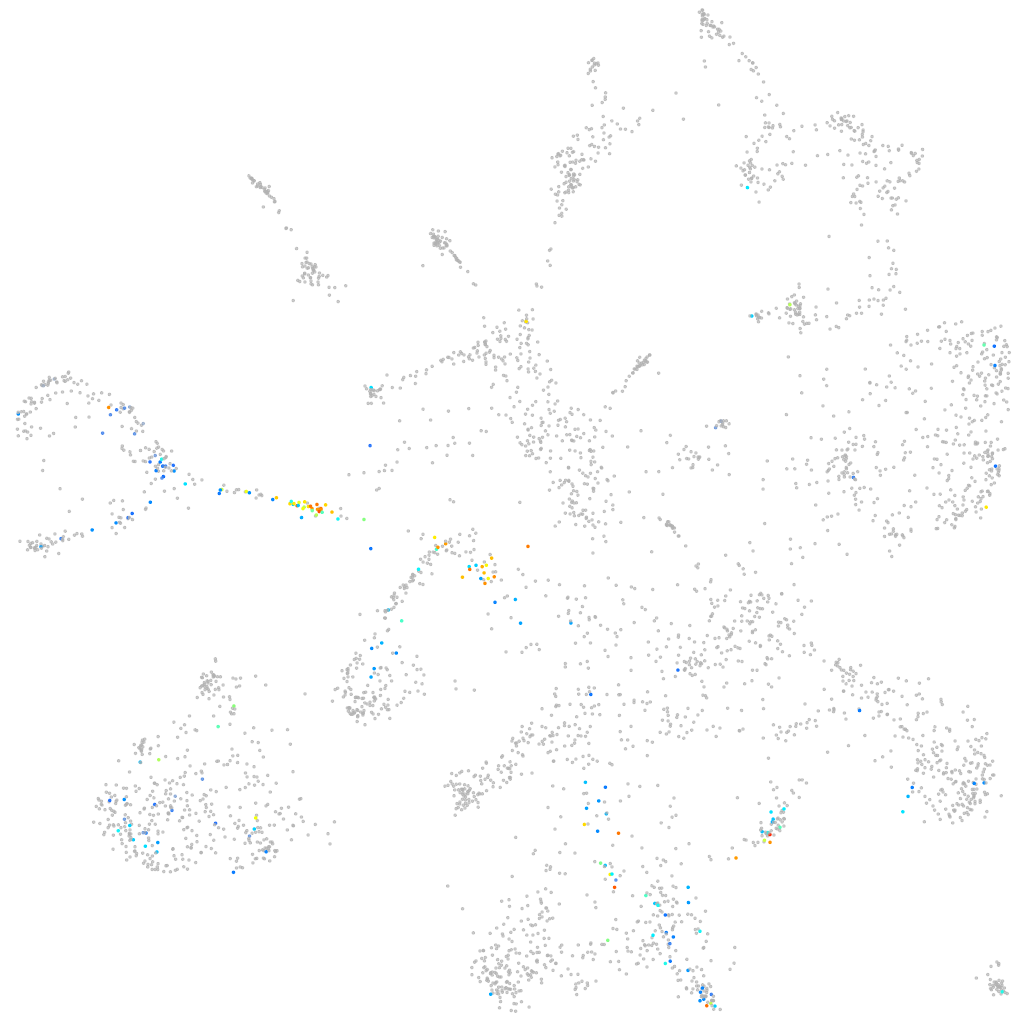
"syncoilin, intermediate filament protein"
ZFIN 
Other cell groups


all cells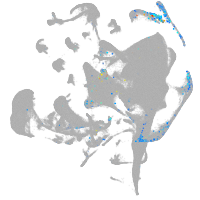 |
blastomeres |
PGCs |
endoderm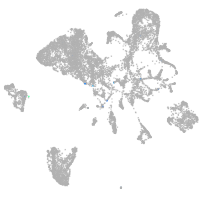 |
axial mesoderm |
muscle 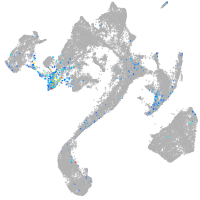 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis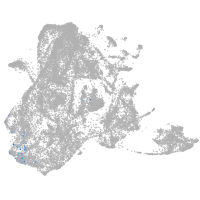 |
periderm |
fin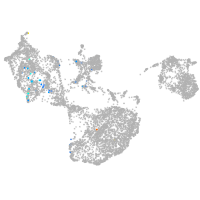 |
ionocytes / mucous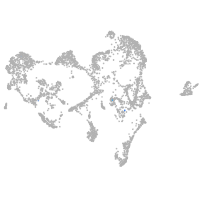 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tacr2 | 0.289 | mdka | -0.134 |
| tnfaip6 | 0.288 | krt8 | -0.121 |
| barx1 | 0.252 | krt18a.1 | -0.112 |
| sh3bgrl2 | 0.243 | akap12b | -0.098 |
| inhbab | 0.236 | marcksl1a | -0.089 |
| adh8a | 0.223 | itga5 | -0.080 |
| tac3b | 0.219 | podxl | -0.080 |
| tspan15 | 0.216 | fstl1a | -0.079 |
| zgc:153704 | 0.207 | si:dkey-238c7.16 | -0.077 |
| hhip | 0.203 | si:dkey-261h17.1 | -0.076 |
| bcan | 0.201 | CABZ01072614.1 | -0.076 |
| mamdc2a | 0.200 | cfd | -0.075 |
| wnt4a | 0.198 | jam2b | -0.074 |
| si:ch211-105c13.3 | 0.196 | notch3 | -0.072 |
| emilin2a | 0.188 | krt18b | -0.072 |
| mylkb | 0.185 | c1qtnf6a | -0.071 |
| alpl | 0.183 | hic1 | -0.070 |
| angptl5 | 0.172 | ftr82 | -0.069 |
| scube2 | 0.171 | lsp1 | -0.069 |
| rrad | 0.171 | mmp16a | -0.069 |
| isl2b | 0.171 | marcksl1b | -0.069 |
| ckba | 0.167 | si:ch73-281n10.2 | -0.067 |
| lmod1b | 0.164 | twist1a | -0.067 |
| gucy1a1 | 0.163 | cd248a | -0.064 |
| foxf2a | 0.162 | krt94 | -0.063 |
| fbxl22 | 0.159 | pdgfrb | -0.063 |
| tbx3b | 0.159 | rps29 | -0.063 |
| hoxb5a | 0.157 | rhag | -0.063 |
| acta2 | 0.156 | nkx2.3 | -0.062 |
| myl9a | 0.155 | ednraa | -0.062 |
| fgf10a | 0.155 | hspb1 | -0.062 |
| htr1d | 0.153 | fabp11a | -0.061 |
| tes | 0.152 | sept15 | -0.060 |
| si:dkeyp-75h12.2 | 0.152 | XLOC-020301 | -0.060 |
| si:ch73-173h19.3 | 0.148 | mab21l1 | -0.059 |