"SPT4 homolog, DSIF elongation factor subunit"
ZFIN 
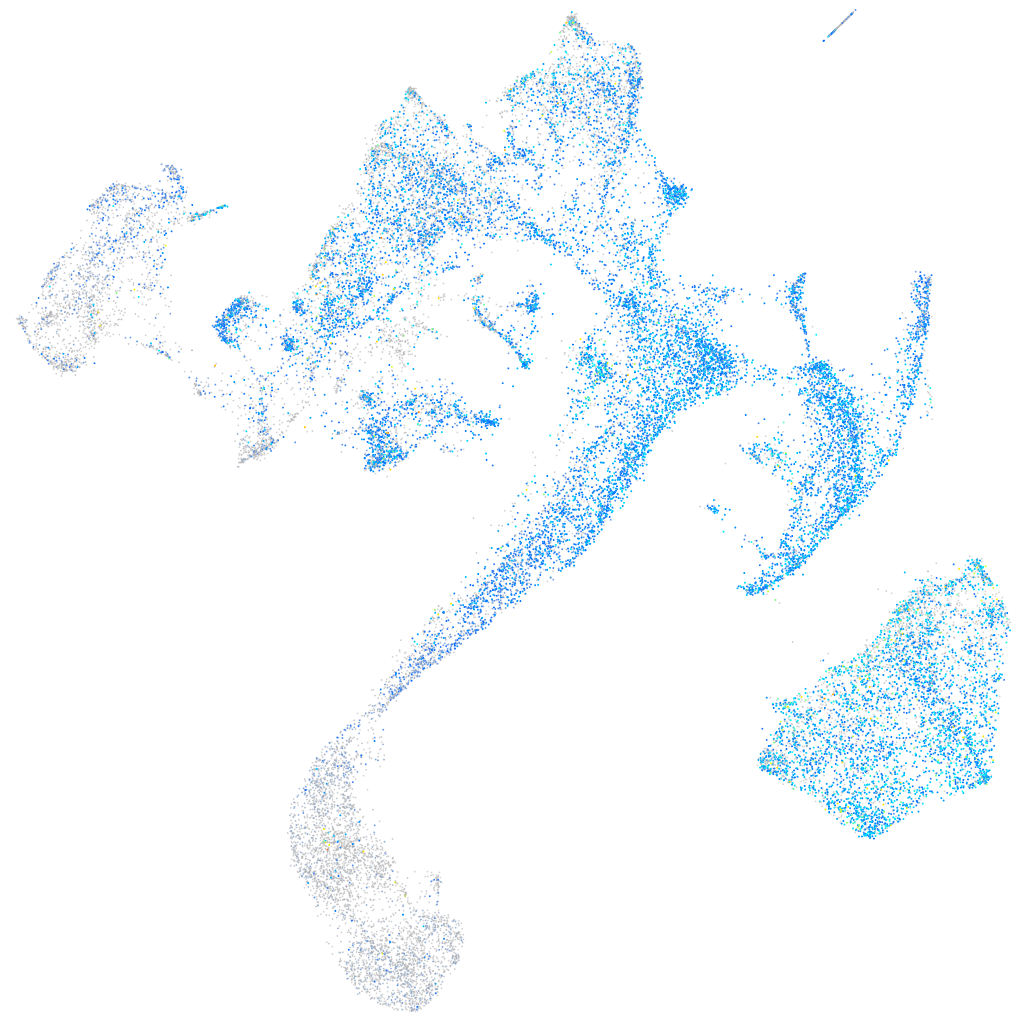
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hnrnpabb | 0.470 | ckma | -0.412 |
| khdrbs1a | 0.462 | ckmb | -0.406 |
| h2afvb | 0.462 | actc1b | -0.405 |
| hnrnpaba | 0.452 | atp2a1 | -0.398 |
| setb | 0.449 | ak1 | -0.397 |
| h3f3d | 0.445 | neb | -0.391 |
| ran | 0.443 | tnnc2 | -0.389 |
| hmga1a | 0.443 | si:ch73-367p23.2 | -0.384 |
| snrpb | 0.436 | actn3a | -0.381 |
| cbx3a | 0.436 | ldb3b | -0.376 |
| rbm8a | 0.435 | gapdh | -0.374 |
| snrpf | 0.433 | myom1a | -0.373 |
| npm1a | 0.433 | pvalb1 | -0.370 |
| hnrnpa0b | 0.432 | eno3 | -0.370 |
| hmgb2b | 0.431 | aldoab | -0.369 |
| snu13b | 0.431 | mylpfa | -0.368 |
| ddx39ab | 0.429 | smyd1a | -0.366 |
| cirbpb | 0.427 | actn3b | -0.366 |
| snrpd2 | 0.422 | si:ch211-266g18.10 | -0.365 |
| snrpd1 | 0.419 | pgam2 | -0.364 |
| syncrip | 0.418 | pvalb2 | -0.363 |
| nop58 | 0.412 | mylz3 | -0.362 |
| cirbpa | 0.412 | tnnt3a | -0.362 |
| hmgb2a | 0.411 | nme2b.2 | -0.361 |
| snrpe | 0.410 | tmod4 | -0.360 |
| ncl | 0.408 | XLOC-025819 | -0.359 |
| srsf3b | 0.408 | acta1b | -0.357 |
| si:ch73-281n10.2 | 0.406 | ank1a | -0.356 |
| nhp2 | 0.405 | prx | -0.352 |
| sap18 | 0.404 | XLOC-001975 | -0.350 |
| ptges3b | 0.403 | tnnt3b | -0.350 |
| h2afva | 0.399 | casq2 | -0.349 |
| nop10 | 0.397 | ttn.2 | -0.349 |
| pabpc1a | 0.397 | hhatla | -0.349 |
| hmgn7 | 0.397 | tpma | -0.347 |