"SGT1 homolog, MIS12 kinetochore complex assembly cochaperone"
ZFIN 
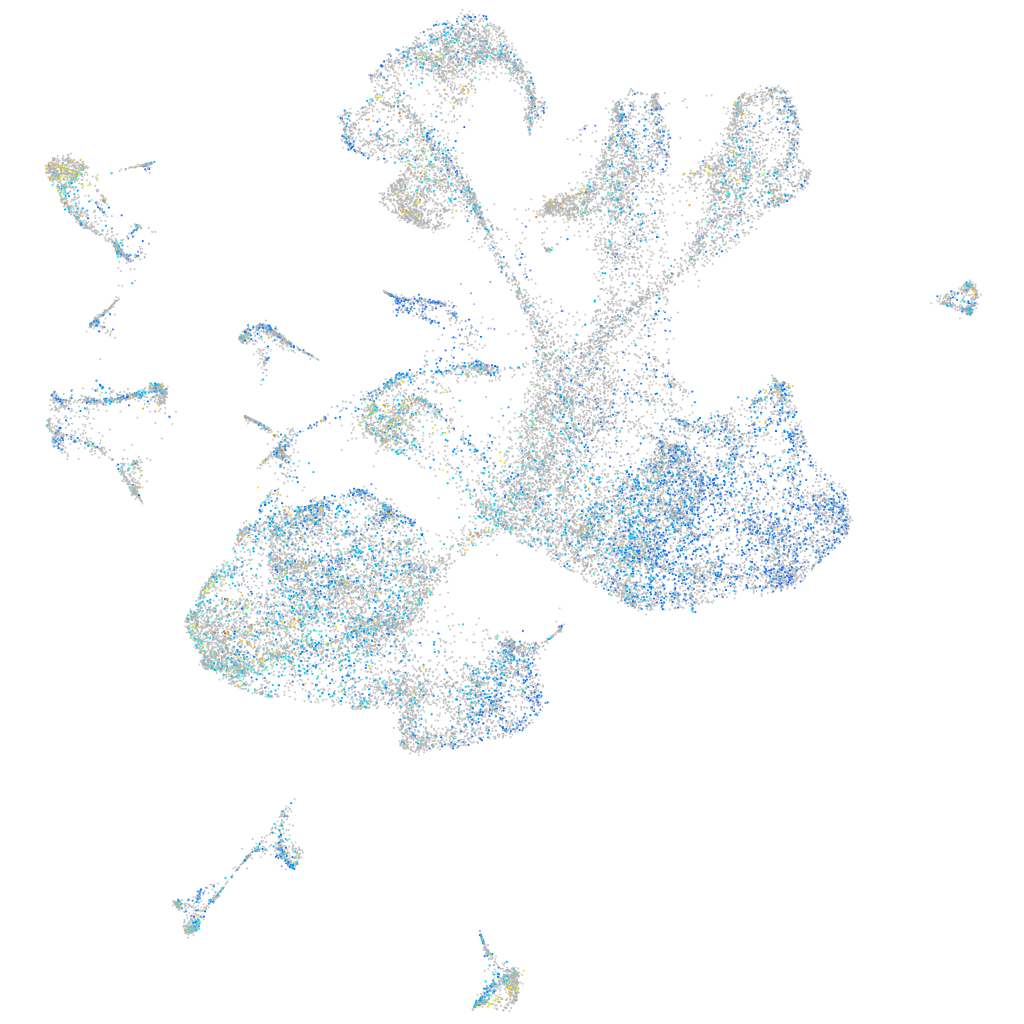
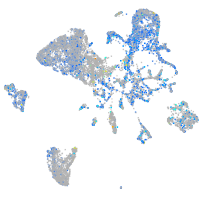
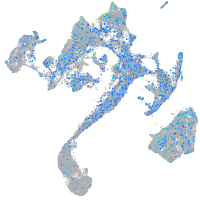
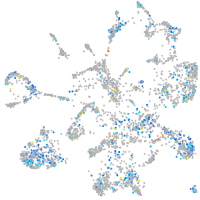
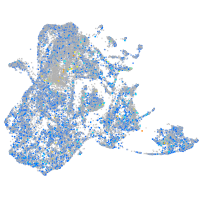
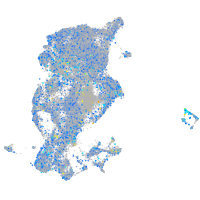
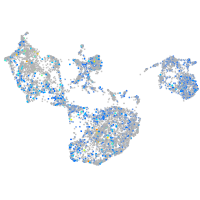
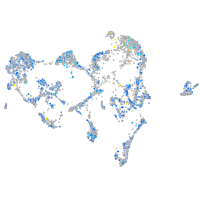
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| fabp7a | 0.102 | elavl3 | -0.105 |
| qki2 | 0.098 | myt1b | -0.089 |
| GCA | 0.093 | myt1a | -0.086 |
| efhd1 | 0.092 | tp53inp2 | -0.080 |
| sept10 | 0.089 | dlb | -0.079 |
| ppap2d | 0.088 | ebf2 | -0.071 |
| si:dkey-204f11.64 | 0.087 | nhlh2 | -0.071 |
| gnai2a | 0.085 | sox11b | -0.070 |
| cspg5b | 0.085 | LOC100537384 | -0.068 |
| cd82a | 0.085 | si:ch73-386h18.1 | -0.068 |
| atp1b4 | 0.083 | rtn1a | -0.065 |
| psat1 | 0.083 | scrt2 | -0.065 |
| phgdh | 0.082 | srrm4 | -0.065 |
| tubb4b | 0.081 | ptmab | -0.064 |
| sox2 | 0.080 | pik3r3b | -0.064 |
| cnn2 | 0.080 | si:ch211-222l21.1 | -0.062 |
| tuba8l | 0.080 | onecut2 | -0.062 |
| sri | 0.079 | LOC798783 | -0.062 |
| id1 | 0.079 | tmsb | -0.062 |
| si:dkey-7j14.6 | 0.079 | insm1a | -0.060 |
| atp1a1b | 0.079 | onecut1 | -0.060 |
| pgrmc1 | 0.079 | zfhx3 | -0.059 |
| rap1b | 0.078 | fstl1a | -0.057 |
| atp1b1a | 0.078 | scrt1a | -0.055 |
| ccng1 | 0.077 | gadd45gb.1 | -0.055 |
| anxa11b | 0.077 | gng2 | -0.055 |
| cd63 | 0.077 | h3f3d | -0.055 |
| psph | 0.076 | cdkn1ca | -0.055 |
| klf6a | 0.076 | stxbp1a | -0.054 |
| hepacama | 0.076 | spsb4a | -0.054 |
| rpz5 | 0.075 | hmgb3a | -0.054 |
| slc1a3b | 0.075 | nova2 | -0.054 |
| anxa13 | 0.075 | lima1a | -0.053 |
| zgc:153867 | 0.074 | elovl4a | -0.053 |
| sept8b | 0.074 | neurod4 | -0.052 |