syntaxin binding protein 4
ZFIN 
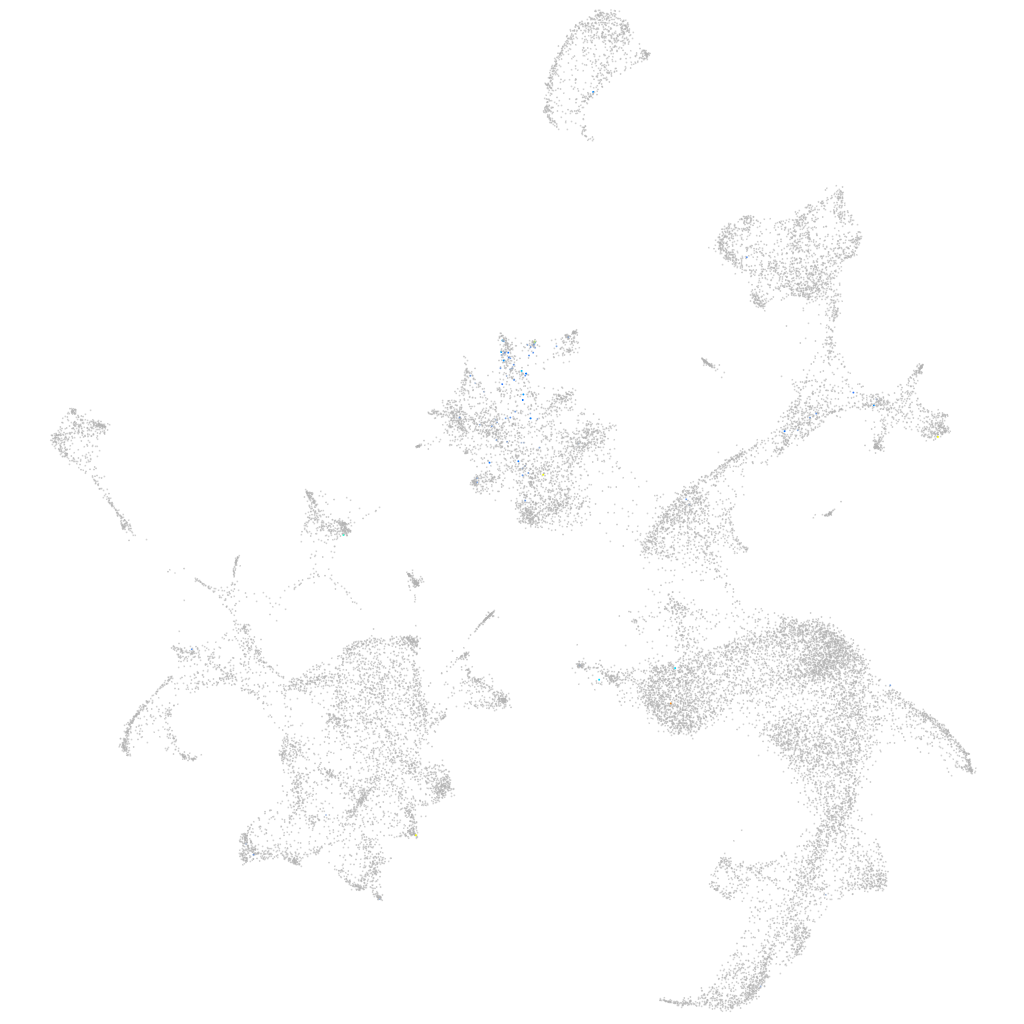
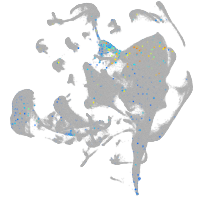
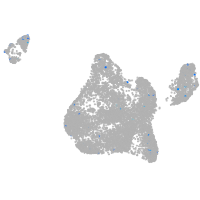
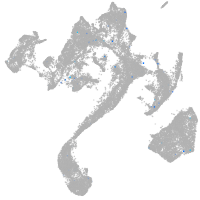
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CR925761.3 | 0.149 | hbbe3 | -0.023 |
| faimb | 0.140 | urod | -0.019 |
| cdk5r2b | 0.139 | ap2m1a | -0.019 |
| neto2a | 0.130 | znfl2a | -0.017 |
| grin2aa | 0.128 | fam210b | -0.017 |
| syt1b | 0.127 | zgc:56095 | -0.017 |
| itga3a | 0.127 | f13a1b | -0.017 |
| BX120006.3 | 0.127 | si:ch211-227m13.1 | -0.016 |
| LOC103910313 | 0.122 | tal1 | -0.016 |
| si:ch1073-155h21.2 | 0.122 | atp1b1a | -0.016 |
| arl3l2 | 0.120 | epb41b | -0.016 |
| pcdh2ab12 | 0.119 | zgc:163057 | -0.015 |
| cdhr1a | 0.118 | TCIM (1 of many) | -0.015 |
| onecut3a | 0.118 | CT030188.1 | -0.015 |
| opn6a | 0.117 | blvrb | -0.015 |
| zgc:109965 | 0.116 | hdr | -0.015 |
| ckmt2a | 0.116 | anxa4 | -0.015 |
| pcdh15b | 0.116 | alad | -0.015 |
| si:dkey-225k4.1 | 0.114 | gata1a | -0.014 |
| saga | 0.114 | she | -0.014 |
| gc2 | 0.113 | cldng | -0.014 |
| tulp1b | 0.111 | tfr1a | -0.014 |
| guca1c | 0.109 | vamp5 | -0.014 |
| rbp4l | 0.109 | blf | -0.014 |
| XLOC-015093 | 0.109 | fech | -0.013 |
| rcvrna | 0.108 | anxa11a | -0.013 |
| pde6c | 0.107 | tspan4b | -0.013 |
| rcvrn2 | 0.106 | blvra | -0.013 |
| hmgcra | 0.106 | rhocb | -0.013 |
| elovl4b | 0.105 | tmem14ca | -0.013 |
| rom1a | 0.102 | b4galt1l | -0.013 |
| si:dkey-17e16.15 | 0.101 | etv2 | -0.013 |
| pde6h | 0.101 | hhex | -0.013 |
| prom1b | 0.100 | ctsa | -0.013 |
| pimr205 | 0.100 | hmbsb | -0.013 |