serine/threonine kinase receptor associated protein
ZFIN 
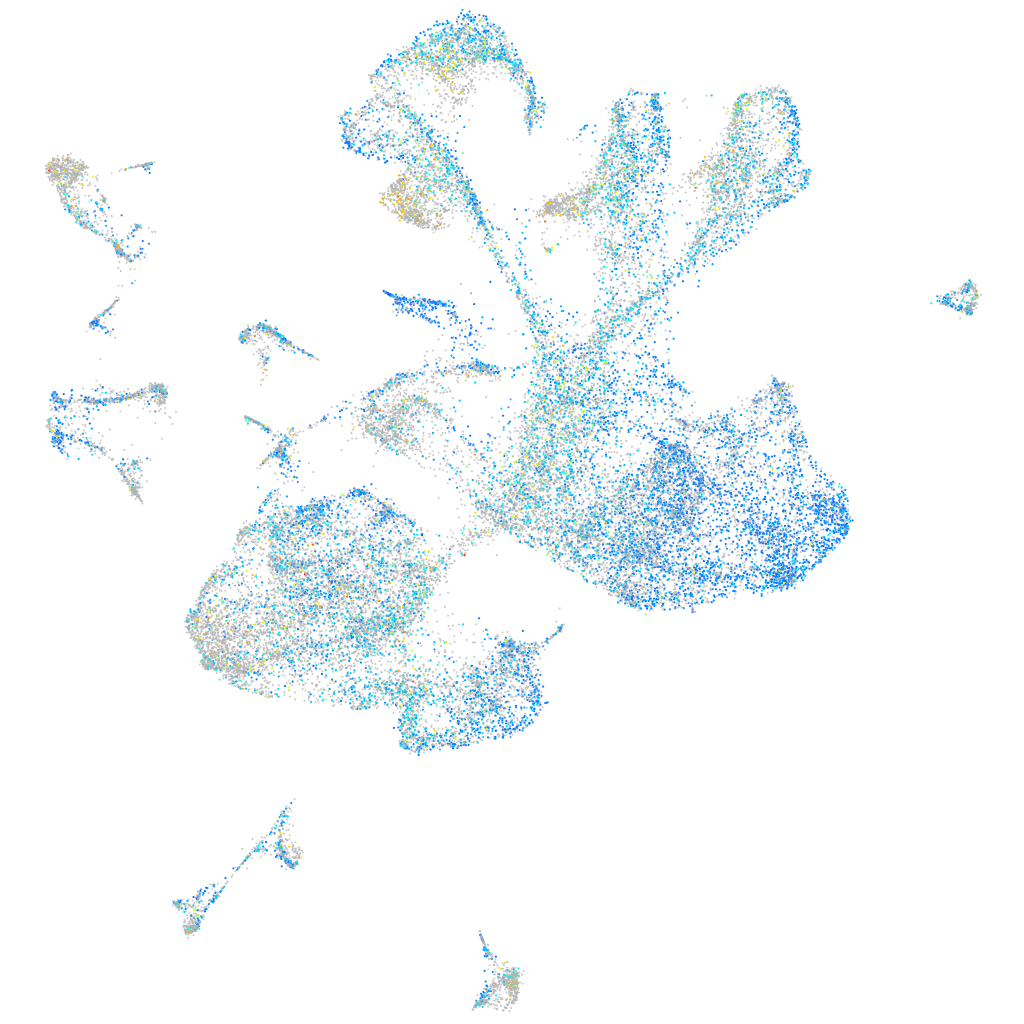
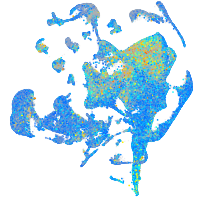
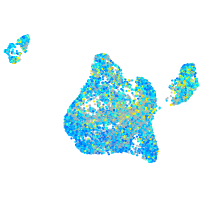
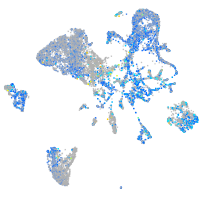
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hnrnpa0l | 0.154 | atp1a1b | -0.114 |
| hnrnpaba | 0.154 | fabp7a | -0.090 |
| khdrbs1a | 0.150 | slc1a2b | -0.090 |
| cirbpb | 0.140 | ptn | -0.088 |
| hsp90ab1 | 0.137 | cx43 | -0.086 |
| eif3ba | 0.135 | slc4a4a | -0.084 |
| hnrnpabb | 0.132 | glula | -0.081 |
| hspa8 | 0.132 | gpr37l1b | -0.078 |
| cct5 | 0.131 | si:ch211-66e2.5 | -0.074 |
| sumo3a | 0.128 | atp1b4 | -0.074 |
| h3f3d | 0.128 | qki2 | -0.073 |
| snrpd2 | 0.128 | efhd1 | -0.072 |
| cct3 | 0.127 | ppap2d | -0.072 |
| slc25a5 | 0.126 | slc3a2a | -0.071 |
| cct2 | 0.125 | mdkb | -0.068 |
| eif3ea | 0.124 | zgc:158463 | -0.067 |
| hnrnpa0b | 0.122 | hepacama | -0.067 |
| ran | 0.122 | si:ch211-251b21.1 | -0.067 |
| cct7 | 0.121 | cox4i2 | -0.066 |
| eef1a1l1 | 0.121 | CR383676.1 | -0.065 |
| ilf2 | 0.120 | mt2 | -0.065 |
| cct4 | 0.120 | cdo1 | -0.064 |
| rpl19 | 0.120 | slc6a11b | -0.064 |
| tcp1 | 0.119 | slc1a3b | -0.063 |
| h2afvb | 0.119 | slc6a9 | -0.062 |
| ybx1 | 0.118 | s1pr1 | -0.061 |
| serbp1a | 0.118 | CU467822.1 | -0.061 |
| eif3f | 0.117 | cd63 | -0.061 |
| hdac1 | 0.117 | zgc:153704 | -0.060 |
| si:ch211-222l21.1 | 0.117 | slc6a1b | -0.060 |
| naca | 0.117 | mfge8a | -0.060 |
| eef1g | 0.117 | pvalb1 | -0.059 |
| tubb2b | 0.117 | cspg5b | -0.058 |
| rps7 | 0.117 | acbd7 | -0.058 |
| ddx39ab | 0.116 | sept8b | -0.058 |