stonin 2
ZFIN 
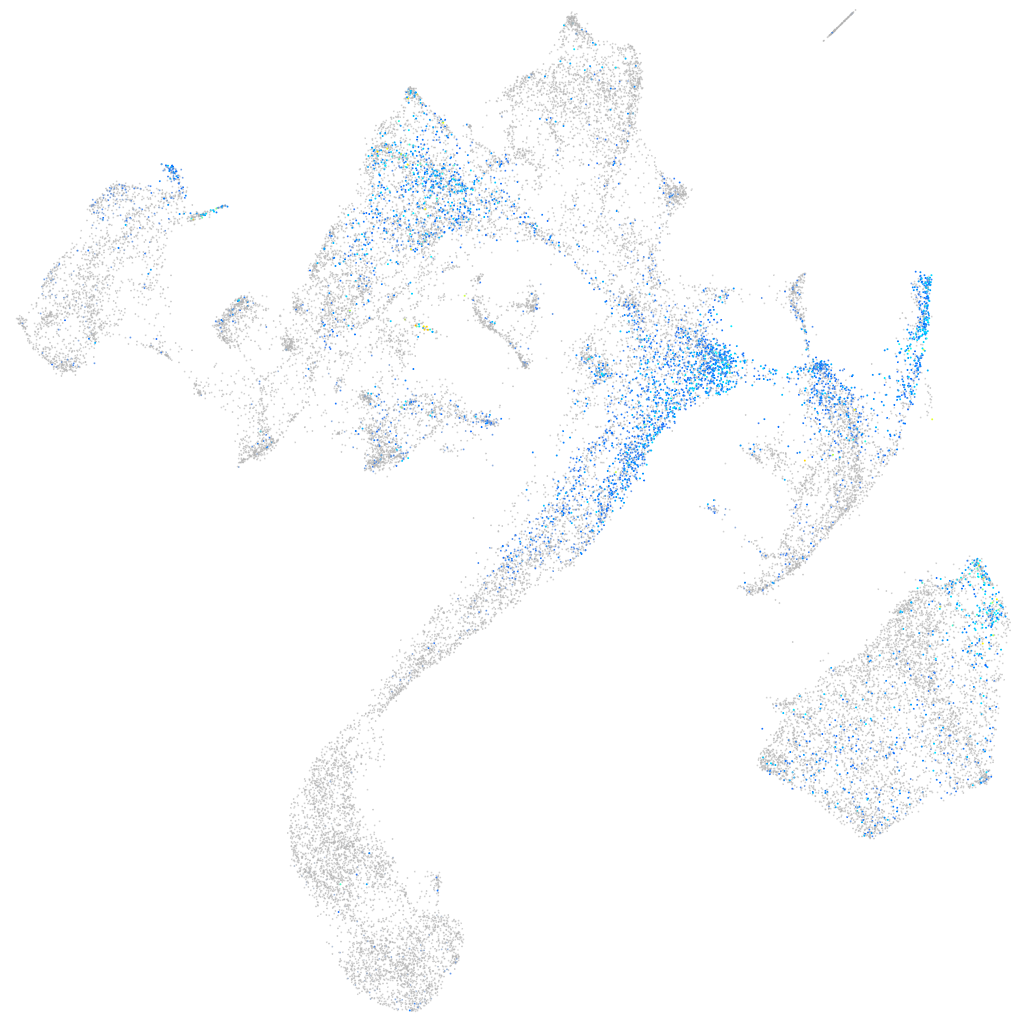
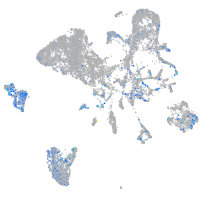
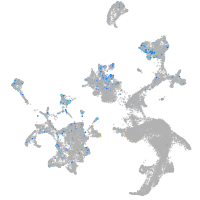
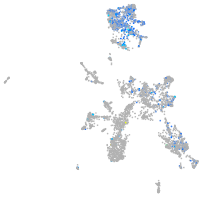
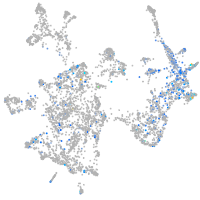
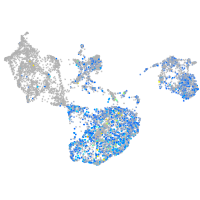
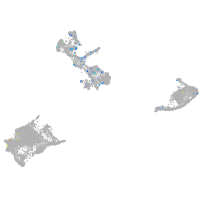
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| XLOC-042229 | 0.319 | ckma | -0.186 |
| ripply1 | 0.291 | ckmb | -0.177 |
| zbtb18 | 0.285 | atp2a1 | -0.173 |
| bmpr1ba | 0.279 | tnnt3a | -0.170 |
| tjp2b | 0.263 | mylpfa | -0.169 |
| meox1 | 0.256 | mylz3 | -0.167 |
| kazald2 | 0.255 | si:ch73-367p23.2 | -0.163 |
| rbm24a | 0.253 | gapdh | -0.163 |
| fn1b | 0.251 | pvalb1 | -0.162 |
| tcf15 | 0.251 | ank1a | -0.160 |
| LOC110438394 | 0.249 | pvalb2 | -0.159 |
| XLOC-009784 | 0.246 | ldb3b | -0.159 |
| dmrt2a | 0.246 | tnnt3b | -0.157 |
| cpn1 | 0.245 | myom1a | -0.155 |
| efemp2a | 0.241 | tnni2a.4 | -0.155 |
| plpp1a | 0.236 | mylpfb | -0.151 |
| XLOC-040516 | 0.235 | ak1 | -0.149 |
| draxin | 0.235 | pgam2 | -0.149 |
| rasgef1ba | 0.232 | prx | -0.149 |
| qkia | 0.229 | myl1 | -0.148 |
| mdka | 0.229 | eno3 | -0.147 |
| fat2 | 0.219 | XLOC-025819 | -0.146 |
| XLOC-029508 | 0.217 | myom2a | -0.146 |
| fstl1a | 0.216 | hhatla | -0.144 |
| sema3ab | 0.216 | cavin4a | -0.144 |
| selenof | 0.207 | casq1b | -0.143 |
| zgc:92429 | 0.206 | XLOC-001975 | -0.143 |
| LOC103910903 | 0.205 | si:ch211-266g18.10 | -0.143 |
| efnb2a | 0.205 | actn3a | -0.142 |
| optc | 0.204 | myhz1.3 | -0.141 |
| nhsl1b | 0.200 | neb | -0.141 |
| net1 | 0.198 | si:ch211-255p10.3 | -0.141 |
| fam49a | 0.196 | myoz1b | -0.140 |
| emp2 | 0.194 | XLOC-005350 | -0.139 |
| cdh2 | 0.191 | eef2l2 | -0.139 |