stonin 2
ZFIN 
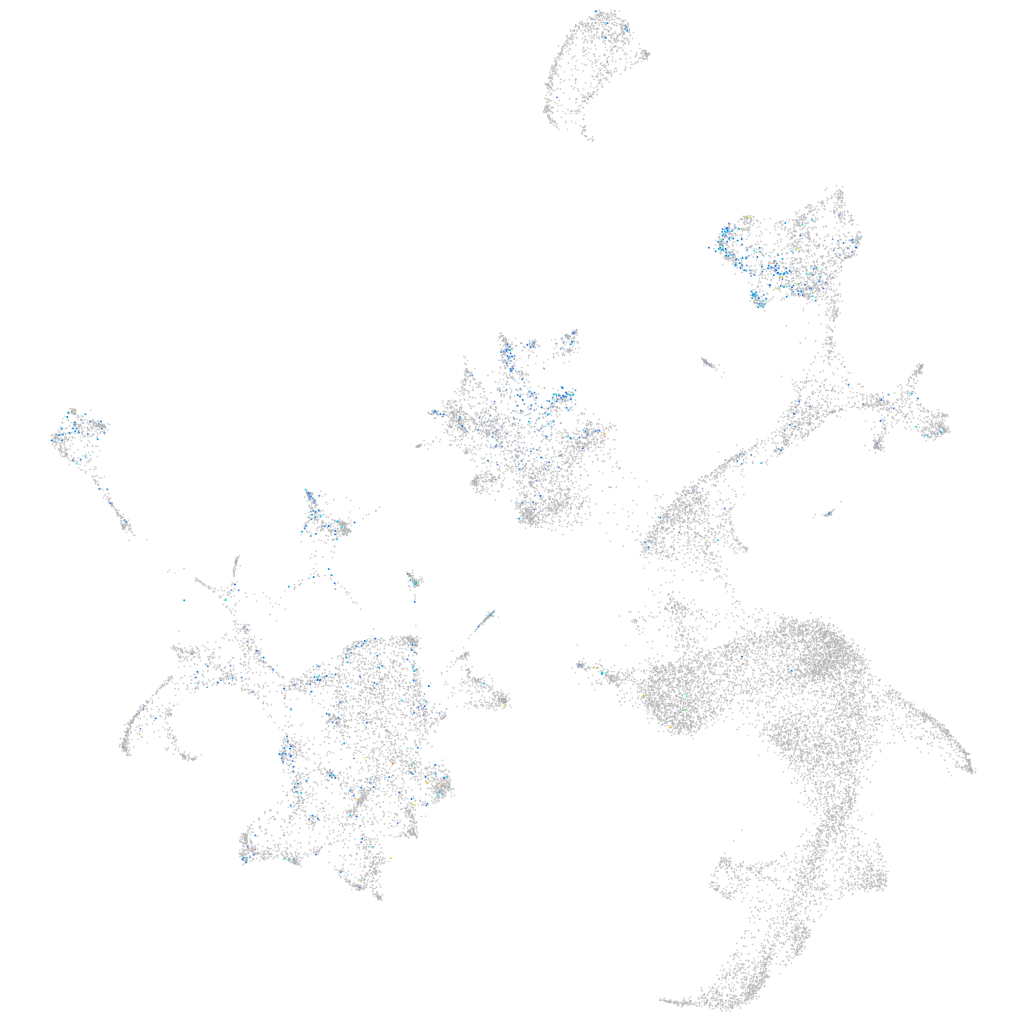
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| slc7a7 | 0.159 | hbae1.1 | -0.108 |
| slc3a2a | 0.154 | hbae3 | -0.101 |
| si:zfos-1069f5.1 | 0.154 | hbae1.3 | -0.100 |
| mpp1 | 0.151 | hbbe1.3 | -0.100 |
| ccl34b.1 | 0.147 | cahz | -0.098 |
| cmklr1 | 0.143 | hbbe2 | -0.097 |
| ctsc | 0.143 | hemgn | -0.095 |
| sdc4 | 0.141 | hbbe1.1 | -0.095 |
| havcr1 | 0.141 | alas2 | -0.092 |
| abcc5 | 0.140 | slc4a1a | -0.092 |
| mpeg1.1 | 0.138 | tspo | -0.090 |
| si:ch73-158p21.3 | 0.137 | hbbe1.2 | -0.089 |
| ccl35.1 | 0.133 | nt5c2l1 | -0.087 |
| slc43a2a | 0.133 | zgc:163057 | -0.086 |
| rnaset2 | 0.132 | blvrb | -0.083 |
| lgals3bpb | 0.131 | epb41b | -0.081 |
| tnfsf12 | 0.129 | nmt1b | -0.079 |
| slc40a1 | 0.128 | hbbe3 | -0.077 |
| ctsba | 0.128 | prdx2 | -0.075 |
| npc2 | 0.128 | fth1a | -0.075 |
| CU499330.1 | 0.127 | plac8l1 | -0.074 |
| glula | 0.126 | tmod4 | -0.073 |
| lgals9l1 | 0.126 | zgc:56095 | -0.072 |
| ctsl.1 | 0.125 | rfesd | -0.070 |
| c1qc | 0.125 | creg1 | -0.070 |
| bzw2 | 0.125 | znfl2a | -0.067 |
| si:ch211-106a19.1 | 0.122 | hbae5 | -0.067 |
| abhd12 | 0.122 | si:ch211-207c6.2 | -0.067 |
| marco | 0.121 | uros | -0.066 |
| psap | 0.121 | klf1 | -0.066 |
| si:dkey-5n18.1 | 0.121 | si:ch211-250g4.3 | -0.065 |
| slc43a2b | 0.121 | tfr1a | -0.063 |
| ctsla | 0.120 | sptb | -0.063 |
| itgae.1 | 0.120 | hdr | -0.062 |
| si:ch211-283g2.2 | 0.119 | ucp3 | -0.061 |