stromal interaction molecule 1a
ZFIN 
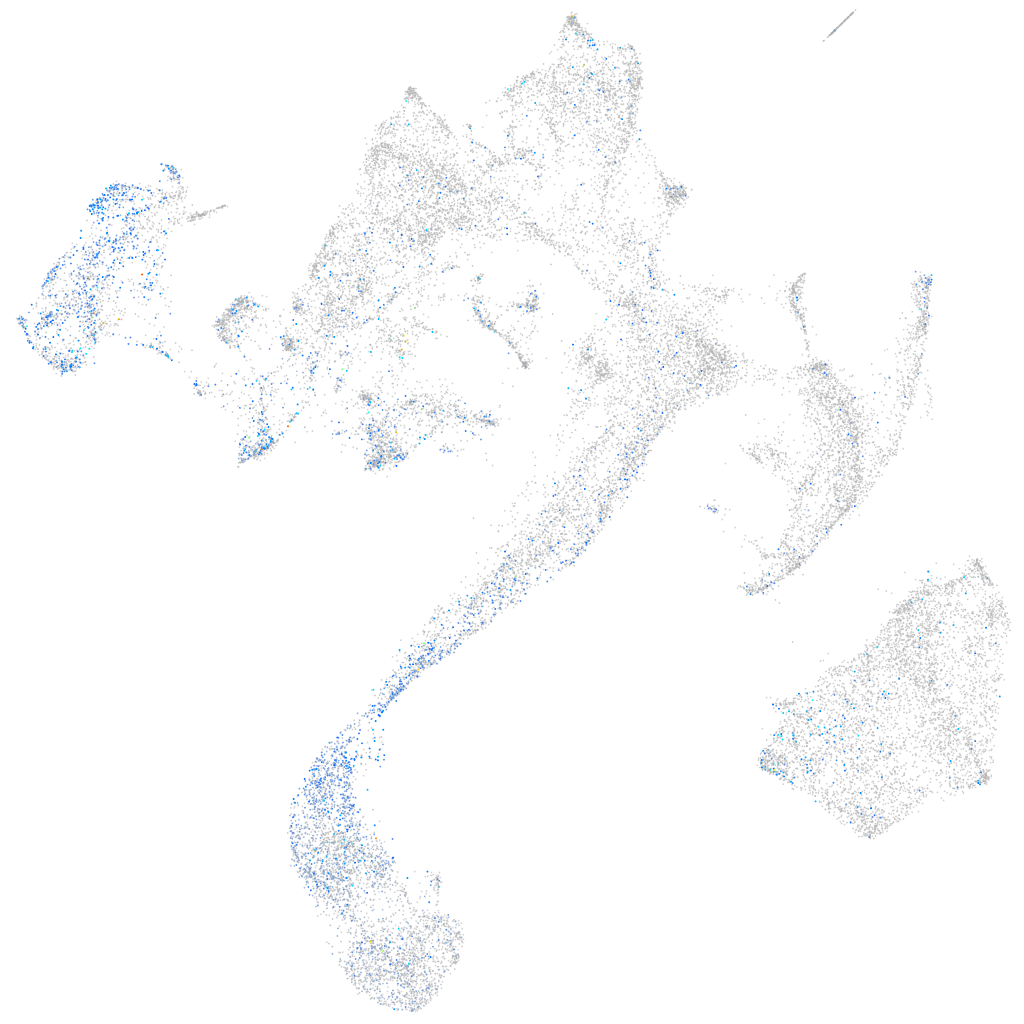
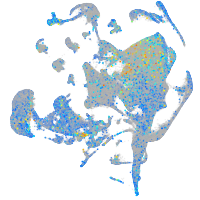
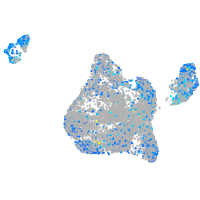
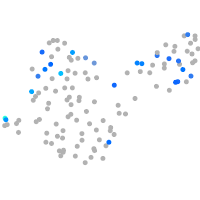
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| casq2 | 0.315 | hsp90ab1 | -0.255 |
| CABZ01072309.1 | 0.310 | pabpc1a | -0.231 |
| srl | 0.307 | h3f3d | -0.224 |
| cav3 | 0.298 | hmgb2b | -0.223 |
| atp2a1 | 0.296 | ran | -0.222 |
| cavin4b | 0.296 | si:ch73-1a9.3 | -0.219 |
| cavin4a | 0.295 | h2afvb | -0.218 |
| aldoab | 0.284 | cirbpb | -0.209 |
| desma | 0.281 | hmgb2a | -0.209 |
| zgc:158296 | 0.281 | khdrbs1a | -0.208 |
| ldb3b | 0.280 | si:ch73-281n10.2 | -0.208 |
| neb | 0.280 | si:ch211-222l21.1 | -0.205 |
| trdn | 0.280 | ubc | -0.205 |
| gapdh | 0.279 | hnrnpaba | -0.203 |
| tmem38a | 0.279 | hmgn2 | -0.200 |
| pgam2 | 0.279 | ptmab | -0.196 |
| si:ch211-266g18.10 | 0.277 | hnrnpabb | -0.192 |
| ckmb | 0.276 | hmga1a | -0.190 |
| prx | 0.273 | hnrnpa0b | -0.190 |
| ckma | 0.273 | cirbpa | -0.189 |
| ank1a | 0.272 | actb2 | -0.186 |
| myom1a | 0.270 | fthl27 | -0.185 |
| tmem182a | 0.270 | hmgn7 | -0.185 |
| nmrk2 | 0.268 | eef1a1l1 | -0.185 |
| ak1 | 0.268 | setb | -0.182 |
| actn3b | 0.268 | cbx3a | -0.182 |
| xirp2a | 0.268 | syncrip | -0.181 |
| ttn.2 | 0.267 | anp32b | -0.179 |
| eno3 | 0.267 | tuba8l4 | -0.178 |
| stac3 | 0.266 | anp32a | -0.177 |
| klhl43 | 0.265 | si:ch211-288g17.3 | -0.175 |
| CABZ01078594.1 | 0.264 | cfl1 | -0.174 |
| sar1ab | 0.263 | h2afva | -0.173 |
| sptb | 0.263 | cx43.4 | -0.172 |
| pmp22b | 0.263 | snrpf | -0.172 |